8HVX
 
 | |
8HVY
 
 | |
8HVZ
 
 | |
8HVW
 
 | |
8IG4
 
 | |
8IG5
 
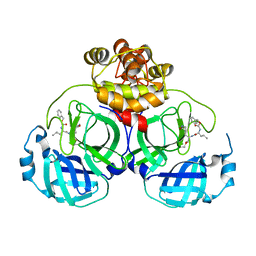 | | Crystal structure of SARS main protease in complex with GC376 | | 分子名称: | 3C-like proteinase nsp5, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | 著者 | Lin, C, Zhang, J, Li, J. | | 登録日 | 2023-02-20 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Structural Basis for Coronaviral Main Proteases Inhibition by the 3CLpro Inhibitor GC376.
J.Mol.Biol., 436, 2024
|
|
8IG6
 
 | |
8IG9
 
 | |
8IGA
 
 | |
8IGB
 
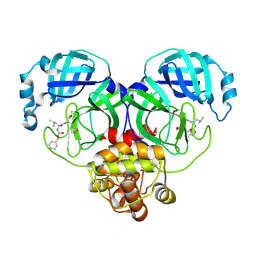 | |
5LFA
 
 | | Crystal structure of iron-sulfur cluster containing bacterial (6-4) photolyase PhrB - Y424F mutant with impaired DNA repair activity | | 分子名称: | (6-4) photolyase, 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Kwiatkowski, D, Zhang, F, Krauss, N, Lamparter, T, Scheerer, P. | | 登録日 | 2016-06-30 | | 公開日 | 2017-01-11 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structures of Bacterial (6-4) Photolyase Mutants with Impaired DNA Repair Activity.
Photochem. Photobiol., 93, 2017
|
|
6CF8
 
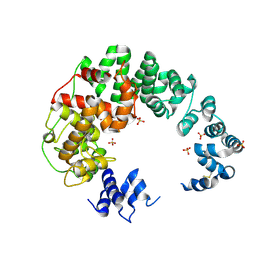 | |
4ZYL
 
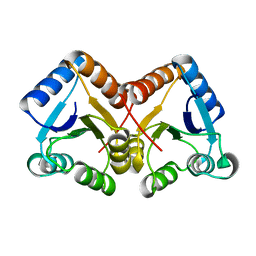 | |
6UVB
 
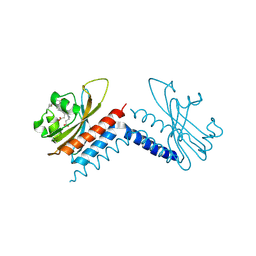 | |
6UV8
 
 | |
2I1T
 
 | |
6DD6
 
 | |
6DD7
 
 | |
3L1L
 
 | |
2OK3
 
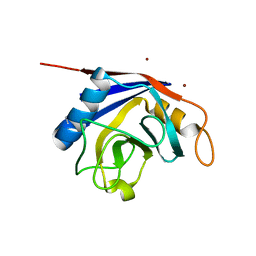 | | X-ray structure of human cyclophilin J at 2.0 angstrom | | 分子名称: | NICKEL (II) ION, Peptidyl-prolyl cis-trans isomerase-like 3 | | 著者 | Xia, Z. | | 登録日 | 2007-01-15 | | 公開日 | 2008-01-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Targeting Cyclophilin J, a novel peptidyl-prolyl isomerase, can induce cellular G1/S arrest and repress the growth of Hepatocellular carcinoma
To be Published
|
|
2OJU
 
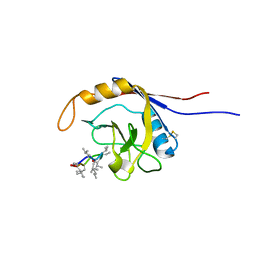 | |
5TUW
 
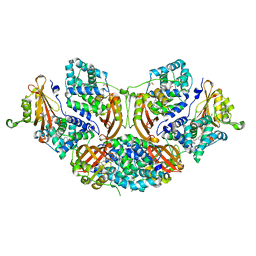 | | Crystal structure of Orange Carotenoid Protein with partial loss of 3'OH Echinenone chromophore | | 分子名称: | (3'R)-3'-hydroxy-beta,beta-caroten-4-one, GLYCEROL, Orange carotenoid-binding protein | | 著者 | Yang, X, Bandara, S, Ren, Z. | | 登録日 | 2016-11-07 | | 公開日 | 2017-06-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.302 Å) | | 主引用文献 | Photoactivation mechanism of a carotenoid-based photoreceptor.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5TV0
 
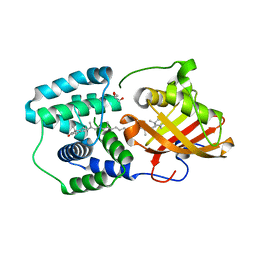 | | crystal structure and light induced structural changes in orange carotenoid protein bound with 3 'OH echinenone | | 分子名称: | (3'R)-3'-hydroxy-beta,beta-caroten-4-one, GLYCEROL, Orange carotenoid-binding protein | | 著者 | Yang, X, Bandara, S, Ren, Z. | | 登録日 | 2016-11-07 | | 公開日 | 2017-06-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.648 Å) | | 主引用文献 | Photoactivation mechanism of a carotenoid-based photoreceptor.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5TUX
 
 | | crystal structure and light induced structural changes in orange carotenoid protein bound with echinenone | | 分子名称: | GLYCEROL, Orange carotenoid-binding protein, beta,beta-caroten-4-one | | 著者 | Yang, X, Bandara, S, Ren, Z. | | 登録日 | 2016-11-07 | | 公開日 | 2017-06-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Photoactivation mechanism of a carotenoid-based photoreceptor.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7LAM
 
 | | Crystal structure of Campylobacter jejuni Cj0843c lytic transglycosylase in complex with N,N',N''-triacetylchitotriose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CITRIC ACID, ... | | 著者 | van den Akker, F, Kumar, V. | | 登録日 | 2021-01-06 | | 公開日 | 2021-04-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Turnover Chemistry and Structural Characterization of the Cj0843c Lytic Transglycosylase of Campylobacter jejuni .
Biochemistry, 60, 2021
|
|
