7LJW
 
 | | X-ray radiation damage series on Thaumatin at 277K, multi-conformer model, dataset 1 | | 分子名称: | L(+)-TARTARIC ACID, Thaumatin I | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-01 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LOR
 
 | | X-ray radiation damage series on Lysozyme at 277K, multi-conformer model, dataset 3 | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-10 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LPM
 
 | | X-ray radiation damage series on Lysozyme at 277K, multi-conformer model, crystal 2 | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-12 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LK6
 
 | | X-ray radiation damage series on Thaumatin at 277K, multi-conformer model, dataset 4 | | 分子名称: | L(+)-TARTARIC ACID, Thaumatin I | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-01 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LLP
 
 | | X-ray radiation damage series on Lysozyme at 277K, crystal structure, dataset 1 | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-04 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LP6
 
 | | X-ray radiation damage series on Lysozyme at 277K, multi-conformer model, dataset 2 (merged) | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-11 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LN8
 
 | | X-ray radiation damage series on Lysozyme at 277K, crystal structure, dataset 3 | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-06 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LQ8
 
 | | X-ray radiation damage series on Proteinase K at 277K, multi-conformer model, dataset 3 | | 分子名称: | CALCIUM ION, Proteinase K, SULFATE ION | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-13 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LJZ
 
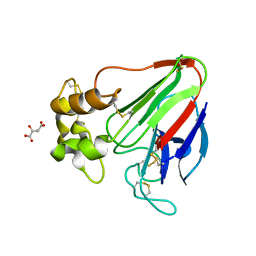 | | X-ray radiation damage series on Thaumatin at 277K, multi-conformer model, dataset 2 | | 分子名称: | L(+)-TARTARIC ACID, Thaumatin I | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-01 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LNB
 
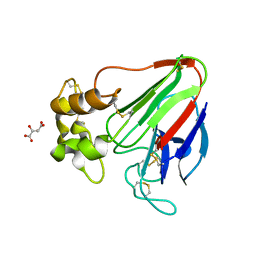 | | X-ray radiation damage series on Thaumatin at 277K, multi-conformer model, dataset 2 (merged) | | 分子名称: | L(+)-TARTARIC ACID, Thaumatin I | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-06 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LPT
 
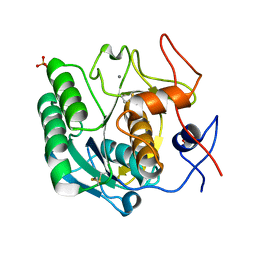 | | X-ray radiation damage series on Proteinase K at 277K, crystal structure, dataset 4 | | 分子名称: | CALCIUM ION, Proteinase K, SULFATE ION | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-12 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LQC
 
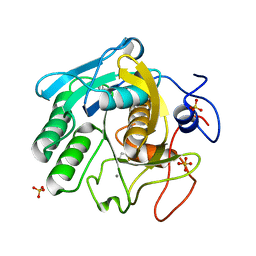 | | X-ray radiation damage series on Proteinase K at 277K, multi-conformer model, dataset 4 (merged) | | 分子名称: | CALCIUM ION, Proteinase K, SULFATE ION | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-13 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.02 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LTD
 
 | | X-ray radiation damage series on Proteinase K at 100K, crystal structure, dataset 1 | | 分子名称: | CALCIUM ION, NITRATE ION, Proteinase K | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-19 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LTI
 
 | | X-ray radiation damage series on Proteinase K at 100K, crystal structure, dataset 2 | | 分子名称: | CALCIUM ION, NITRATE ION, Proteinase K | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-19 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (0.91 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LTV
 
 | | X-ray radiation damage series on Proteinase K at 100K, crystal structure, dataset 3 | | 分子名称: | CALCIUM ION, NITRATE ION, Proteinase K | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-20 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (0.95 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LU0
 
 | | X-ray radiation damage series on Proteinase K at 100K, crystal structure, dataset 4 | | 分子名称: | CALCIUM ION, NITRATE ION, Proteinase K | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-20 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.01 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LU1
 
 | | X-ray radiation damage series on Proteinase K at 100K, crystal structure, dataset 5 | | 分子名称: | CALCIUM ION, NITRATE ION, Proteinase K | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-20 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.06 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LU2
 
 | | X-ray radiation damage series on Proteinase K at 100K, crystal structure, dataset 6 | | 分子名称: | CALCIUM ION, NITRATE ION, Proteinase K | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-20 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.11 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LU3
 
 | | X-ray radiation damage series on Proteinase K at 100K, crystal structure, dataset 7 | | 分子名称: | CALCIUM ION, NITRATE ION, Proteinase K | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-20 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4HEO
 
 | |
5UGI
 
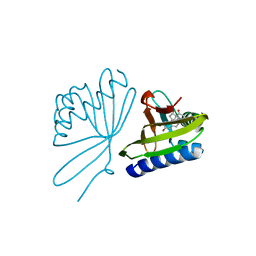 | |
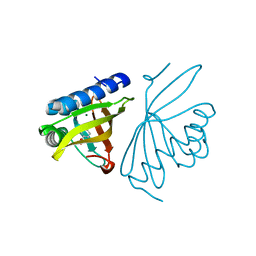
6C1J
 
 | | Crystal Structure of Ketosteroid Isomerase Y32F/Y57F/D40N mutant from Pseudomonas Putida (pKSI) bound to 3,4-dinitrophenol | | 分子名称: | 3,4-dinitrophenol, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Yabukarski, F, Pinney, M.M, Herschlag, D. | | 登録日 | 2018-01-04 | | 公開日 | 2018-07-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.063 Å) | | 主引用文献 | Structural Coupling Throughout the Active Site Hydrogen Bond Networks of Ketosteroid Isomerase and Photoactive Yellow Protein.
J. Am. Chem. Soc., 140, 2018
|
|
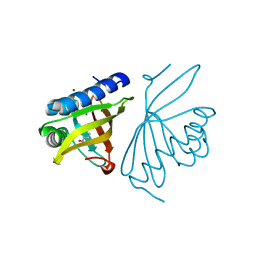
6C17
 
 | | Crystal Structure of Ketosteroid Isomerase D40N mutant from Pseudomonas Putida (pKSI) bound to 3,4-dinitrophenol | | 分子名称: | 3,4-dinitrophenol, MAGNESIUM ION, Steroid Delta-isomerase | | 著者 | Yabukarski, F, Pinney, M, Herschlag, D. | | 登録日 | 2018-01-04 | | 公開日 | 2018-07-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Structural Coupling Throughout the Active Site Hydrogen Bond Networks of Ketosteroid Isomerase and Photoactive Yellow Protein.
J. Am. Chem. Soc., 140, 2018
|
|
6C1X
 
 | | Crystal Structure of Ketosteroid Isomerase D40N/D103N mutant from Pseudomonas Putida (pKSI) bound to 3,4-dinitrophenol | | 分子名称: | 3,4-dinitrophenol, MAGNESIUM ION, Steroid Delta-isomerase | | 著者 | Yabukarski, F, Pinney, M.M, Herschlag, D. | | 登録日 | 2018-01-05 | | 公開日 | 2018-07-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Structural Coupling Throughout the Active Site Hydrogen Bond Networks of Ketosteroid Isomerase and Photoactive Yellow Protein.
J. Am. Chem. Soc., 140, 2018
|
|
6P44
 
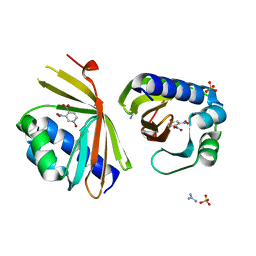 | | Crystal Structure of Ketosteroid Isomerase D38N mutant from Mycobacterium hassiacum (mhKSI) bound to 3,4-dinitrophenol | | 分子名称: | 3,4-dinitrophenol, GUANIDINE, SULFATE ION, ... | | 著者 | Yabukarski, F, Doukov, T, Pinney, M, Herschlag, D. | | 登録日 | 2019-05-25 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.251 Å) | | 主引用文献 | Parallel molecular mechanisms for enzyme temperature adaptation.
Science, 371, 2021
|
|
