6HAG
 
 | |
6HTJ
 
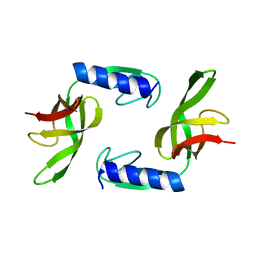 | |
6EWS
 
 | | Solution Structure of Rhabdopeptide NRPS Docking Domain Kj12A-NDD | | 分子名称: | NRPS Kj12A-NDD | | 著者 | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | 登録日 | 2017-11-06 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
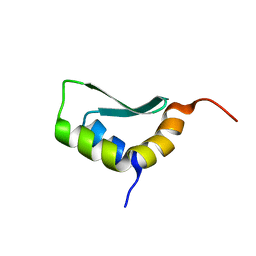
6EWV
 
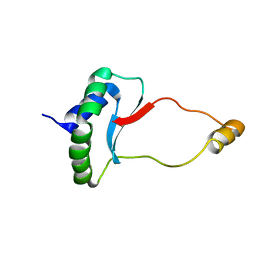 | | Solution Structure of Docking Domain Complex of RXP NRPS: Kj12C NDD - Kj12B CDD | | 分子名称: | NRPS Kj12C-NDD, NRPS Kj12B-CDD | | 著者 | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | 登録日 | 2017-11-06 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
6EWT
 
 | | Solution Structure of Rhabdopeptide NRPS Docking Domain Kj12B-NDD | | 分子名称: | NRPS Kj12B-NDD | | 著者 | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | 登録日 | 2017-11-06 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
4YMH
 
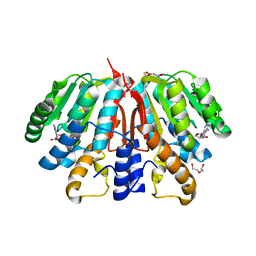 | | Crystal structure of SAH-bound Podospora anserina methyltransferase PaMTH1 | | 分子名称: | DI(HYDROXYETHYL)ETHER, Putative SAM-dependent O-methyltranferase, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Kudlinzki, D, Linhard, V.L, Chatterjee, D, Saxena, K, Sreeramulu, S, Schwalbe, H. | | 登録日 | 2015-03-06 | | 公開日 | 2015-05-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.876 Å) | | 主引用文献 | Structure and Biophysical Characterization of the S-Adenosylmethionine-dependent O-Methyltransferase PaMTH1, a Putative Enzyme Accumulating during Senescence of Podospora anserina.
J.Biol.Chem., 290, 2015
|
|
1B75
 
 | |
1C2X
 
 | |
1C2W
 
 | |
4I68
 
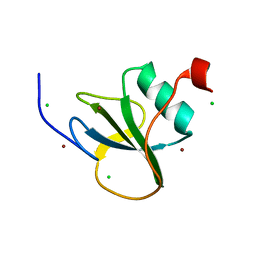 | |
4I67
 
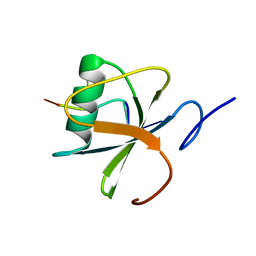 | |
4I69
 
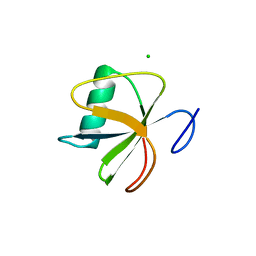 | |
2KMJ
 
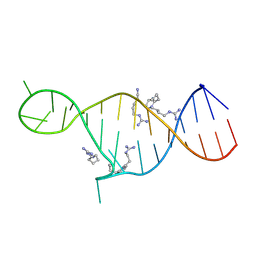 | | High resolution NMR solution structure of a complex of HIV-2 TAR RNA and a synthetic tripeptide in a 1:2 stoichiometry | | 分子名称: | Pyrimidinylpeptide, RNA (28-MER) | | 著者 | Ferner, J, Suhartono, M, Breitung, S, Jonker, H.R.A, Hennig, M, Woehnert, J, Goebel, M, Schwalbe, H. | | 登録日 | 2009-07-30 | | 公開日 | 2009-08-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structures of HIV TAR RNA-ligand complexes reveal higher binding stoichiometries.
Chembiochem, 10, 2009
|
|
1MNX
 
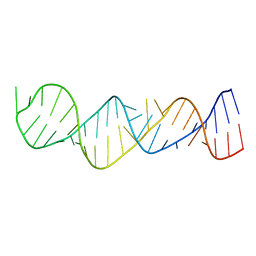 | |
2LVL
 
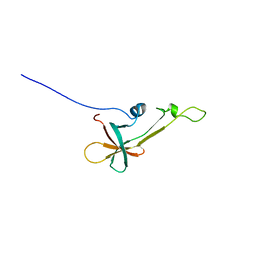 | | NMR Structure the lantibiotic immunity protein SpaI | | 分子名称: | SpaI | | 著者 | Christ, N, Bochmann, S, Gottstein, D, Duchardt-Ferner, E, Hellmich, U.A, Duesterhus, S, Koetter, P, Guentert, P, Entian, K, Woehnert, J. | | 登録日 | 2012-07-06 | | 公開日 | 2012-08-29 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The First Structure of a Lantibiotic Immunity Protein, SpaI from Bacillus subtilis, Reveals a Novel Fold.
J.Biol.Chem., 287, 2012
|
|
2LCQ
 
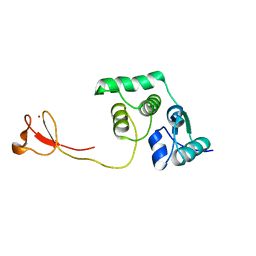 | | Solution structure of the endonuclease Nob1 from P.horikoshii | | 分子名称: | Putative toxin VapC6, ZINC ION | | 著者 | Veith, T, Martin, R, Wurm, J.P, Weis, B, Duchardt-Ferner, E, Safferthal, C, Hennig, R, Mirus, O, Bohnsack, M.T, Woehnert, J, Schleiff, E. | | 登録日 | 2011-05-05 | | 公開日 | 2011-12-14 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and functional analysis of the archaeal endonuclease Nob1.
Nucleic Acids Res., 40, 2012
|
|
2N2E
 
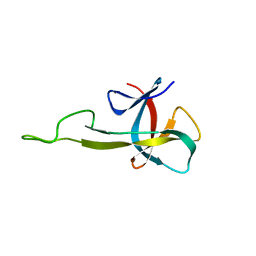 | | NMR solution structure of the C-terminal domain of NisI, a lipoprotein from Lactococcus lactis which confers immunity against nisin | | 分子名称: | Nisin immunity protein | | 著者 | Hacker, C, Christ, N.A, Korn, S, Duchardt-Ferner, E, Hellmich, U.A, Duesterhus, S, Koetter, P, Entian, K, Woehnert, J. | | 登録日 | 2015-05-08 | | 公開日 | 2015-10-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Solution Structure of the Lantibiotic Immunity Protein NisI and Its Interactions with Nisin.
J.Biol.Chem., 290, 2015
|
|
2N32
 
 | | NMR solution structure of the N-terminal domain of NisI, a lipoprotein from Lactococcus lactis which confers immunity against nisin | | 分子名称: | Nisin immunity protein | | 著者 | Hacker, C, Christ, N.A, Korn, S, Duchardt-Ferner, E, Hellmich, U.A, Duesterhus, S, Koetter, P, Entian, K, Woehnert, J. | | 登録日 | 2015-05-21 | | 公開日 | 2015-10-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Solution Structure of the Lantibiotic Immunity Protein NisI and Its Interactions with Nisin.
J.Biol.Chem., 290, 2015
|
|
