8ETG
 
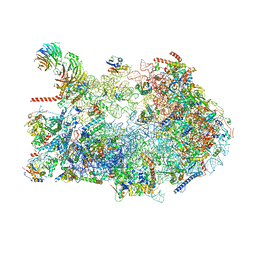 | | Fkbp39 associated 60S nascent ribosome State 3 | | 分子名称: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | 著者 | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | 登録日 | 2022-10-17 | | 公開日 | 2022-11-30 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
2G6U
 
 | |
1E59
 
 | |
7VRD
 
 | |
7V67
 
 | |
1WC8
 
 | | The crystal structure of mouse bet3p | | 分子名称: | MYRISTIC ACID, TRAFFICKING PROTEIN PARTICLE COMPLEX SUBUNIT3 | | 著者 | Kim, Y.-G, Sacher, M, Oh, B.-H. | | 登録日 | 2004-11-10 | | 公開日 | 2004-12-13 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure of Bet3 Reveals a Novel Mechanism for Golgi Localization of Tethering Factor Trapp
Nat.Struct.Mol.Biol., 12, 2005
|
|
1WC9
 
 | | The crystal structure of truncated mouse bet3p | | 分子名称: | GLYCEROL, MYRISTIC ACID, TRAFFICKING PROTEIN PARTICLE COMPLEX SUBUNIT3 | | 著者 | Kim, Y.-G, Lee, H.-S, Sacher, M, Oh, B.-H. | | 登録日 | 2004-11-10 | | 公開日 | 2004-12-13 | | 最終更新日 | 2015-04-15 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal Structure of Bet3 Reveals a Novel Mechanism for Golgi Localization of Tethering Factor Trapp
Nat.Struct.Mol.Biol., 12, 2005
|
|
