4LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | 著者 | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | 登録日 | 1991-01-25 | | 公開日 | 1992-07-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
4V45
 
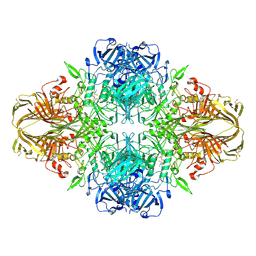 | | E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE | | 分子名称: | 2-deoxy-2-fluoro-beta-D-galactopyranose, Beta-Galactosidase, MAGNESIUM ION, ... | | 著者 | Juers, D.H, McCarter, J.D, Withers, S.G, Matthews, B.W. | | 登録日 | 2001-09-13 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | A Structural View of the Action of Escherichia Coli (Lacz) Beta-Galactosidase
Biochemistry, 40, 2001
|
|
4V44
 
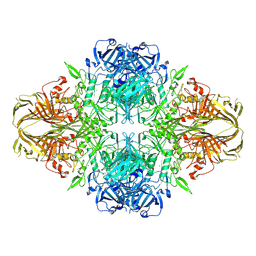 | | E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE | | 分子名称: | 2-deoxy-2-fluoro-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Beta-Galactosidase, MAGNESIUM ION, ... | | 著者 | Juers, D.H, McCarter, J.D, Withers, S.G, Matthews, B.W. | | 登録日 | 2001-09-13 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | A Structural View of the Action of Escherichia Coli (Lacz) Beta-Galactosidase
Biochemistry, 40, 2001
|
|
1TMN
 
 | |
3LZM
 
 | |
6CRO
 
 | |
3F8V
 
 | |
3FA0
 
 | |
3FAD
 
 | |
3ORC
 
 | |
3LVB
 
 | | Crystal structure of the Ferredoxin:NADP+ reductase from maize root at 1.7 angstroms - Test Set Withheld | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin-NADP reductase | | 著者 | Faber, H.R, Karplus, P.A, Aliverti, A, Ferioli, C, Spinola, M. | | 登録日 | 2010-02-19 | | 公開日 | 2010-03-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Biochemical and crystallographic characterization of ferredoxin-NADP(+) reductase from nonphotosynthetic tissues
Biochemistry, 40, 2001
|
|
1L60
 
 | |
1L56
 
 | |
3C83
 
 | |
3C81
 
 | |
3C8S
 
 | |
3DBK
 
 | |
3CDO
 
 | |
3CDR
 
 | |
3CDQ
 
 | |
3C8Q
 
 | |
3CDT
 
 | |
3CDV
 
 | |
3C80
 
 | | T4 Lysozyme mutant R96Y at room temperature | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | 著者 | Mooers, B.H.M. | | 登録日 | 2008-02-08 | | 公開日 | 2009-02-17 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Contributions of all 20 amino acids at site 96 to the stability and structure of T4 lysozyme.
Protein Sci., 18, 2009
|
|
3C8R
 
 | |
