4GKG
 
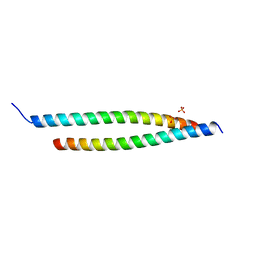 | | Crystal structure of the S-Helix Linker | | 分子名称: | C4-dicarboxylate transport sensor protein dctB, PHOSPHATE ION | | 著者 | Liu, J.W, Lu, D, Sun, Y.J, Wen, J, Yang, Y, Yang, J.G, Wei, X.L, Zhang, X.D, Wang, Y.P. | | 登録日 | 2012-08-11 | | 公開日 | 2013-08-28 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.695 Å) | | 主引用文献 | Crystal structure of the S-Helix Linker
To be Published
|
|
8JML
 
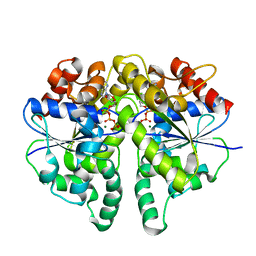 | | Structure of Helicobacter pylori Soj protein mutant, D41A | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, SpoOJ regulator (Soj) | | 著者 | Wu, C.T, Chu, C.H, Sun, Y.J. | | 登録日 | 2023-06-05 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-07-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Insights into the molecular mechanism of ParABS system in chromosome partition by HpParA and HpParB.
Nucleic Acids Res., 52, 2024
|
|
8JMK
 
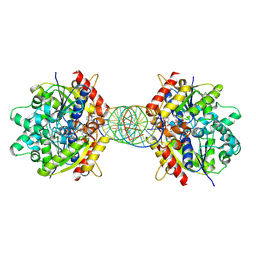 | | Structure of Helicobacter pylori Soj mutant, D41A bound to DNA | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*AP*GP*GP*GP*TP*GP*TP*TP*CP*CP*AP*CP*GP*TP*GP*AP*AP*AP*CP*AP*GP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*GP*TP*TP*TP*CP*AP*CP*GP*TP*GP*GP*AP*AP*CP*AP*CP*CP*CP*T)-3'), ... | | 著者 | Wu, C.T, Chu, C.H, Sun, Y.J. | | 登録日 | 2023-06-05 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-07-24 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Insights into the molecular mechanism of ParABS system in chromosome partition by HpParA and HpParB.
Nucleic Acids Res., 52, 2024
|
|
8JMJ
 
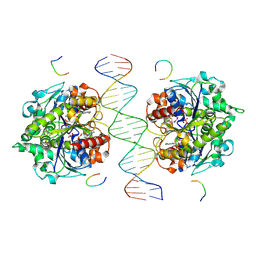 | | Structure of Helicobacter pylori Soj-DNA-Spo0J complex | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*AP*GP*GP*GP*TP*GP*TP*TP*CP*CP*AP*CP*GP*TP*GP*AP*AP*AP*CP*AP*GP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*GP*TP*TP*TP*CP*AP*CP*GP*TP*GP*GP*AP*AP*CP*AP*CP*CP*CP*T)-3'), ... | | 著者 | Wu, C.T, Chu, C.H, Sun, Y.J. | | 登録日 | 2023-06-05 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-07-24 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | Insights into the molecular mechanism of ParABS system in chromosome partition by HpParA and HpParB.
Nucleic Acids Res., 52, 2024
|
|
6LX0
 
 | |
7YJW
 
 | |
7DUT
 
 | | Structure of Sulfolobus solfataricus SegA protein | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SOJ protein (Soj) | | 著者 | Yen, C.Y, Lin, M.G, Hsiao, C.D, Sun, Y.J. | | 登録日 | 2021-01-11 | | 公開日 | 2021-12-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Chromosome segregation in Archaea: SegA- and SegB-DNA complex structures provide insights into segrosome assembly.
Nucleic Acids Res., 49, 2021
|
|
7DUV
 
 | | Structure of Sulfolobus solfataricus SegB protein | | 分子名称: | SULFATE ION, SegB | | 著者 | Yen, C.Y, Lin, M.G, Sun, Y.J, Hsiao, C.D. | | 登録日 | 2021-01-11 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Chromosome segregation in Archaea: SegA- and SegB-DNA complex structures provide insights into segrosome assembly.
Nucleic Acids Res., 49, 2021
|
|
7DV2
 
 | | Structure of Sulfolobus solfataricus SegB-DNA complex | | 分子名称: | DNA (5'-D(P*AP*CP*GP*TP*AP*GP*AP*AP*GP*AP*GP*TP*CP*TP*AP*GP*AP*CP*TP*G)-3'), DNA (5'-D(P*CP*AP*GP*TP*CP*TP*AP*GP*AP*CP*TP*CP*TP*TP*CP*TP*AP*CP*GP*TP*A)-3'), SegB | | 著者 | Yen, C.Y, Lin, M.G, Sun, Y.J, Hsiao, C.D. | | 登録日 | 2021-01-12 | | 公開日 | 2021-12-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Chromosome segregation in Archaea: SegA- and SegB-DNA complex structures provide insights into segrosome assembly.
Nucleic Acids Res., 49, 2021
|
|
7DV3
 
 | | Structure of Sulfolobus solfataricus SegA-AMPPNP protein | | 分子名称: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SOJ protein (Soj) | | 著者 | Yen, C.Y, Lin, M.G, Wu, C.T, Hsiao, C.D, Sun, Y.J. | | 登録日 | 2021-01-12 | | 公開日 | 2021-12-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Chromosome segregation in Archaea: SegA- and SegB-DNA complex structures provide insights into segrosome assembly.
Nucleic Acids Res., 49, 2021
|
|
7DWR
 
 | | Structure of Sulfolobus solfataricus SegA-ADP complex bound to DNA | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*AP*GP*GP*GP*TP*GP*TP*TP*CP*CP*AP*CP*GP*TP*GP*AP*AP*AP*CP*AP*GP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*GP*TP*TP*TP*CP*AP*CP*GP*TP*GP*GP*AP*AP*CP*AP*CP*CP*CP*T)-3'), ... | | 著者 | Yen, C.Y, Lin, M.G, Hsiao, C.D, Sun, Y.J. | | 登録日 | 2021-01-17 | | 公開日 | 2021-12-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Chromosome segregation in Archaea: SegA- and SegB-DNA complex structures provide insights into segrosome assembly.
Nucleic Acids Res., 49, 2021
|
|
7JL6
 
 | |
1KQR
 
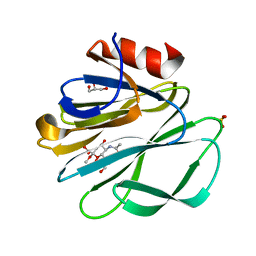 | | Crystal Structure of the Rhesus Rotavirus VP4 Sialic Acid Binding Domain in Complex with 2-O-methyl-alpha-D-N-acetyl neuraminic acid | | 分子名称: | 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, GLYCEROL, SULFATE ION, ... | | 著者 | Dormitzer, P.R, Sun, Z.-Y.J, Wagner, G, Harrison, S.C. | | 登録日 | 2002-01-07 | | 公開日 | 2002-03-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | The Rhesus Rotavirus VP4 Sialic Acid Binding Domain has a Galectin Fold with a
Novel Carbohydrate Binding Site
Embo J., 21, 2002
|
|
1KRI
 
 | |
2AEN
 
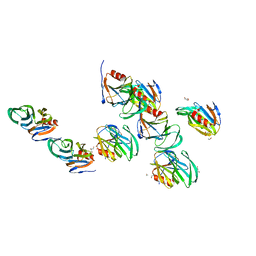 | | Crystal structure of the rotavirus strain DS-1 VP8* core | | 分子名称: | ETHANOL, GLYCEROL, Outer capsid protein VP4, ... | | 著者 | Monnier, N, Higo-Moriguchi, K, Sun, Z.-Y.J, Prasad, B.V.V, Taniguchi, K, Dormitzer, P.R. | | 登録日 | 2005-07-22 | | 公開日 | 2006-02-07 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.604 Å) | | 主引用文献 | High-resolution molecular and antigen structure of the VP8*
core of a sialic acid-independent human rotavirus strain
J.Virol., 80, 2006
|
|
2AIV
 
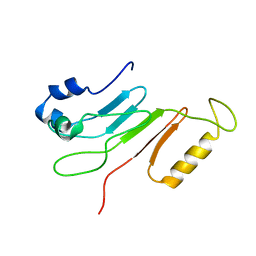 | | Multiple conformations in the ligand-binding site of the yeast nuclear pore targeting domain of NUP116P | | 分子名称: | fragment of Nucleoporin NUP116/NSP116 | | 著者 | Robinson, M.A, Park, S, Sun, Z.-Y.J, Silver, P, Wagner, G, Hogle, J. | | 登録日 | 2005-08-01 | | 公開日 | 2005-08-16 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Multiple Conformations in the Ligand-binding Site of the Yeast Nuclear Pore-targeting Domain of Nup116p
J.Biol.Chem., 280, 2005
|
|
1Z9E
 
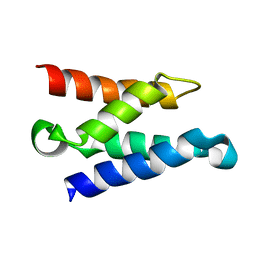 | | Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75 | | 分子名称: | PC4 and SFRS1 interacting protein 2 | | 著者 | Cherepanov, P, Sun, Z.-Y.J, Rahman, S, Maertens, G, Wagner, G, Engelman, A. | | 登録日 | 2005-04-01 | | 公開日 | 2005-05-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75
Nat.Struct.Mol.Biol., 12, 2005
|
|
1WDN
 
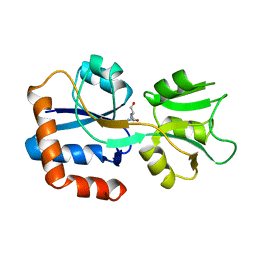 | | GLUTAMINE-BINDING PROTEIN | | 分子名称: | GLUTAMINE, GLUTAMINE BINDING PROTEIN | | 著者 | Sun, Y.-J, Rose, J, Wang, B.-C, Hsiao, C.-D. | | 登録日 | 1997-05-17 | | 公開日 | 1998-05-06 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | The structure of glutamine-binding protein complexed with glutamine at 1.94 A resolution: comparisons with other amino acid binding proteins.
J.Mol.Biol., 278, 1998
|
|
1GSU
 
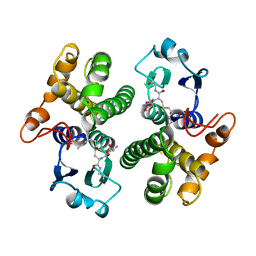 | | AN AVIAN CLASS-MU GLUTATHIONE S-TRANSFERASE, CGSTM1-1 AT 1.94 ANGSTROM RESOLUTION | | 分子名称: | CLASS-MU GLUTATHIONE S-TRANSFERASE, S-HEXYLGLUTATHIONE | | 著者 | Sun, Y.-J, Kuan, C, Tam, M.F, Hsiao, C.-D. | | 登録日 | 1997-09-02 | | 公開日 | 1998-03-04 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | The three-dimensional structure of an avian class-mu glutathione S-transferase, cGSTM1-1 at 1.94 A resolution.
J.Mol.Biol., 278, 1998
|
|
1KXI
 
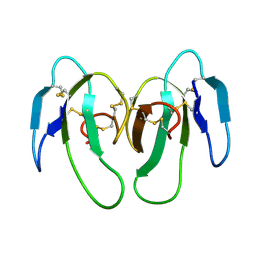 | | STRUCTURE OF CYTOTOXIN HOMOLOG PRECURSOR | | 分子名称: | CARDIOTOXIN V | | 著者 | Sun, Y.-J, Wu, W.-G, Chiang, C.-M, Hsin, A.-Y, Hsiao, C.-D. | | 登録日 | 1996-08-29 | | 公開日 | 1997-04-21 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Crystal structure of cardiotoxin V from Taiwan cobra venom: pH-dependent conformational change and a novel membrane-binding motif identified in the three-finger loops of P-type cardiotoxin.
Biochemistry, 36, 1997
|
|
2PGI
 
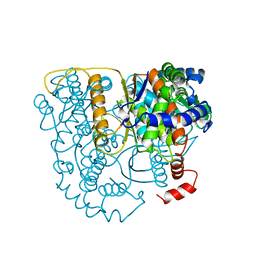 | | THE CRYSTAL STRUCTURE OF PHOSPHOGLUCOSE ISOMERASE-AN ENZYME WITH AUTOCRINE MOTILITY FACTOR ACTIVITY IN TUMOR CELLS | | 分子名称: | PHOSPHOGLUCOSE ISOMERASE | | 著者 | Sun, Y.-J, Chou, C.-C, Chen, W.-S, Meng, M, Hsiao, C.-D. | | 登録日 | 1998-10-27 | | 公開日 | 1999-06-15 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The crystal structure of a multifunctional protein: phosphoglucose isomerase/autocrine motility factor/neuroleukin.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1CI5
 
 | | GLYCAN-FREE MUTANT ADHESION DOMAIN OF HUMAN CD58 (LFA-3) | | 分子名称: | PROTEIN (LYMPHOCYTE FUNCTION-ASSOCIATED ANTIGEN 3(CD58)) | | 著者 | Sun, Z.Y.J, Dotsch, V, Kim, M, Li, J, Reinherz, E.L, Wagner, G. | | 登録日 | 1999-04-07 | | 公開日 | 1999-06-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Functional glycan-free adhesion domain of human cell surface receptor CD58: design, production and NMR studies.
EMBO J., 18, 1999
|
|
5IJ4
 
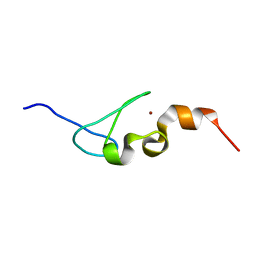 | | Solution structure of AN1-type zinc finger domain from Cuz1 (Cdc48 associated ubiquitin-like/zinc-finger protein-1) | | 分子名称: | CDC48-associated ubiquitin-like/zinc finger protein 1, ZINC ION | | 著者 | Sun, Z.-Y.J, Hanna, J, Wagner, G, Bhanu, M.K, Allan, M, Arthanari, H. | | 登録日 | 2016-03-01 | | 公開日 | 2016-10-05 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of the Cuz1 AN1 Zinc Finger Domain: An Exposed LDFLP Motif Defines a Subfamily of AN1 Proteins.
Plos One, 11, 2016
|
|
1JBJ
 
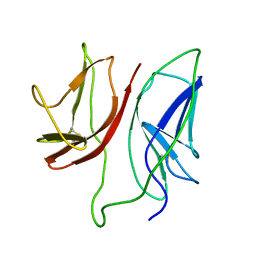 | | CD3 Epsilon and gamma Ectodomain Fragment Complex in Single-Chain Construct | | 分子名称: | CD3 Epsilon and gamma Ectodomain Fragment Complex | | 著者 | Sun, Z.-Y.J, Kim, K.S, Wagner, G, Reinherz, E.L. | | 登録日 | 2001-06-05 | | 公開日 | 2001-12-05 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Mechanisms contributing to T cell receptor signaling and assembly revealed by the solution structure of an ectodomain fragment of the CD3 epsilon gamma heterodimer.
Cell(Cambridge,Mass.), 105, 2001
|
|
1XMW
 
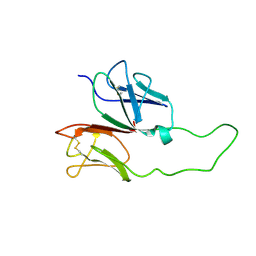 | | CD3 EPSILON AND DELTA ECTODOMAIN FRAGMENT COMPLEX IN SINGLE-CHAIN CONSTRUCT | | 分子名称: | Chimeric CD3 mouse Epsilon and sheep Delta Ectodomain Fragment Complex | | 著者 | Sun, Z.-Y.J, Kim, S.T, Kim, I.C, Fahmy, A, Reinherz, E.L, Wagner, G. | | 登録日 | 2004-10-04 | | 公開日 | 2004-11-30 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the CD3epsilondelta ectodomain and comparison with CD3epsilongamma as a basis for modeling T cell receptor topology and signaling.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
