5GR0
 
 | |
5GR6
 
 | |
5GR3
 
 | |
5GR4
 
 | |
5GQY
 
 | |
5GR1
 
 | |
5GR2
 
 | |
5GR5
 
 | |
5GQZ
 
 | |
8EXL
 
 | | Crystal structure of PI3K-alpha in complex with taselisib | | 分子名称: | 2-methyl-2-(4-{2-[3-methyl-1-(propan-2-yl)-1H-1,2,4-triazol-5-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}-1H-pyrazol-1-yl)propanamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | 著者 | Kiefer, J.R, Eigenbrot, C, Staben, S.T, Hanan, E.J, Wallweber, H.J.A, Ultsch, M, Braun, M.G, Friedman, L.S, Purkey, H.E. | | 登録日 | 2022-10-25 | | 公開日 | 2022-11-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.989 Å) | | 主引用文献 | Discovery of GDC-0077 (Inavolisib), a Highly Selective Inhibitor and Degrader of Mutant PI3K alpha.
J.Med.Chem., 65, 2022
|
|
8EXO
 
 | | Crystal structure of PI3K-alpha in complex with compound 19 | | 分子名称: | 1-{(4S,11aM)-2-[(4R)-2-oxo-4-(propan-2-yl)-1,3-oxazolidin-3-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}-L-prolinamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | 著者 | Kiefer, J.R, Eigenbrot, C, Staben, S.T, Hanan, E.J, Wallweber, H.J.A, Ultsch, M, Braun, M.G, Friedman, L.S, Purkey, H.E. | | 登録日 | 2022-10-25 | | 公開日 | 2022-11-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | Discovery of GDC-0077 (Inavolisib), a Highly Selective Inhibitor and Degrader of Mutant PI3K alpha.
J.Med.Chem., 65, 2022
|
|
8EXU
 
 | | Crystal structure of PI3K-alpha in complex with compound 30 | | 分子名称: | (2S)-2-cyclopropyl-2-({(4S,11aM)-2-[(4S)-2-oxo-4-(trifluoromethyl)-1,3-oxazolidin-3-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}amino)acetamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | 著者 | Kiefer, J.R, Eigenbrot, C, Staben, S.T, Hanan, E.J, Wallweber, H.J.A, Ultsch, M, Braun, M.G, Friedman, L.S, Purkey, H.E. | | 登録日 | 2022-10-25 | | 公開日 | 2022-11-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Discovery of GDC-0077 (Inavolisib), a Highly Selective Inhibitor and Degrader of Mutant PI3K alpha.
J.Med.Chem., 65, 2022
|
|
8EXV
 
 | | Crystal structure of PI3K-alpha in complex with compound 32 | | 分子名称: | N~2~-{(4S,11aP)-2-[(4S)-4-(difluoromethyl)-2-oxo-1,3-oxazolidin-3-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}-L-alaninamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | 著者 | Kiefer, J.R, Eigenbrot, C, Staben, S.T, Hanan, E.J, Wallweber, H.J.A, Ultsch, M, Braun, M.G, Friedman, L.S, Purkey, H.E. | | 登録日 | 2022-10-25 | | 公開日 | 2022-11-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Discovery of GDC-0077 (Inavolisib), a Highly Selective Inhibitor and Degrader of Mutant PI3K alpha.
J.Med.Chem., 65, 2022
|
|
6CHG
 
 | | Crystal structure of the yeast COMPASS catalytic module | | 分子名称: | H3, Histone-lysine N-methyltransferase, H3 lysine-4 specific, ... | | 著者 | Hsu, P.L, Li, H, Zheng, N. | | 登録日 | 2018-02-22 | | 公開日 | 2018-08-22 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.985 Å) | | 主引用文献 | Crystal Structure of the COMPASS H3K4 Methyltransferase Catalytic Module.
Cell, 174, 2018
|
|
4RVK
 
 | | CHK1 kinase domain with diazacarbazole compound 8: N-[3-(6-cyano-9H-pyrrolo[2,3-b:5,4-c']dipyridin-3-yl)phenyl]acetamide | | 分子名称: | N-[3-(6-cyano-9H-pyrrolo[2,3-b:5,4-c']dipyridin-3-yl)phenyl]acetamide, Serine/threonine-protein kinase Chk1 | | 著者 | Wiesmann, C, Wu, P. | | 登録日 | 2014-11-26 | | 公開日 | 2015-06-03 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Mitigation of Acetylcholine Esterase Activity in the 1,7-Diazacarbazole Series of Inhibitors of Checkpoint Kinase 1.
J.Med.Chem., 58, 2015
|
|
4RVL
 
 | | CHK1 kinase domain with diazacarbazole compound 7: 3-(2-hydroxyphenyl)-9H-pyrrolo[2,3-b:5,4-c']dipyridine-6-carbonitrile | | 分子名称: | 3-(2-hydroxyphenyl)-9H-pyrrolo[2,3-b:5,4-c']dipyridine-6-carbonitrile, Serine/threonine-protein kinase Chk1 | | 著者 | Wiesmann, C, Wu, P. | | 登録日 | 2014-11-26 | | 公開日 | 2015-06-03 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Mitigation of Acetylcholine Esterase Activity in the 1,7-Diazacarbazole Series of Inhibitors of Checkpoint Kinase 1.
J.Med.Chem., 58, 2015
|
|
6UH5
 
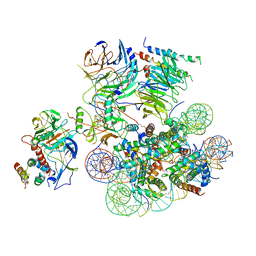 | | Structural basis of COMPASS eCM recognition of the H2Bub nucleosome | | 分子名称: | Bre2, DNA (146-MER), H3 N-terminus, ... | | 著者 | Hsu, P.L, Shi, H, Zheng, N. | | 登録日 | 2019-09-26 | | 公開日 | 2019-11-20 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural Basis of H2B Ubiquitination-Dependent H3K4 Methylation by COMPASS.
Mol.Cell, 76, 2019
|
|
6UGM
 
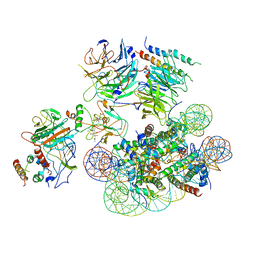 | | Structural basis of COMPASS eCM recognition of an unmodified nucleosome | | 分子名称: | Bre2, DNA (146-MER), H3 N-terminus, ... | | 著者 | Hsu, P.L, Shi, H, Zheng, N. | | 登録日 | 2019-09-26 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural Basis of H2B Ubiquitination-Dependent H3K4 Methylation by COMPASS.
Mol.Cell, 76, 2019
|
|
2HF7
 
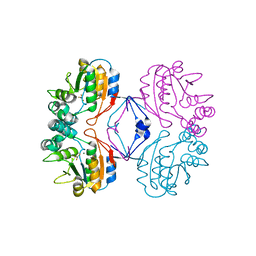 | | Transition State Analogue of AphA class B Acid Phosphatase/Phosphotransferase (Aluminium Fluoride Complex) | | 分子名称: | ALUMINUM FLUORIDE, Class B acid phosphatase, MAGNESIUM ION | | 著者 | Leone, R, Calderone, V, Cappelletti, E, Benvenuti, M, Mangani, S. | | 登録日 | 2006-06-23 | | 公開日 | 2008-03-04 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 |
|
|
2HEG
 
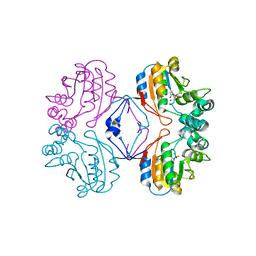 | | Phospho-Aspartyl Intermediate Analogue of Apha class B acid phosphatase/phosphotransferase | | 分子名称: | Class B acid phosphatase, MAGNESIUM ION | | 著者 | Leone, R, Calderone, V, Cappelletti, E, Benvenuti, M, Mangani, S. | | 登録日 | 2006-06-21 | | 公開日 | 2008-03-04 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 |
|
|
5WEV
 
 | |
2G1A
 
 | | Crystal structure of the complex between Apha class B acid phosphatase/phosphotransferase | | 分子名称: | Class B acid phosphatase, MAGNESIUM ION, {[2-(6-AMINO-9H-PURIN-9-YL)ETHOXY]METHYL}PHOSPHONIC ACID | | 著者 | Leone, R, Calderone, V, Cappelletti, E, Benvenuti, M, Mangani, S. | | 登録日 | 2006-02-14 | | 公開日 | 2007-04-24 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of the complex between Apha class B acid phosphatase/phosphotransferase
To be published
|
|
6KLF
 
 | |
3CZ4
 
 | | Native AphA class B acid phosphatase/phosphotransferase from E. coli | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | 著者 | Leone, R, Cappelletti, E, Benvenuti, M, Lentini, G, Thaller, M.C, Mangani, S. | | 登録日 | 2008-04-28 | | 公開日 | 2008-11-11 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural insights into the catalytic mechanism of the bacterial class B phosphatase AphA belonging to the DDDD superfamily of phosphohydrolases.
J.Mol.Biol., 384, 2008
|
|
