4KHP
 
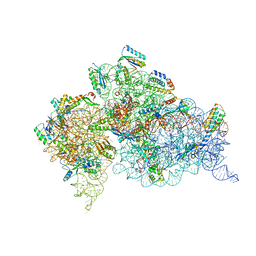 | | Structure of the Thermus thermophilus 30S ribosomal subunit in complex with de-6-MSA-pactamycin | | 分子名称: | 16S Ribosomal RNA, 30S Ribosomal protein S10, 30S Ribosomal protein S11, ... | | 著者 | Tourigny, D.S, Fernandez, I.S, Kelley, A.C, Ramakrishnan, V. | | 登録日 | 2013-05-01 | | 公開日 | 2013-06-05 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Crystal Structure of a Bioactive Pactamycin Analog Bound to the 30S Ribosomal Subunit.
J.Mol.Biol., 425, 2013
|
|
2BE0
 
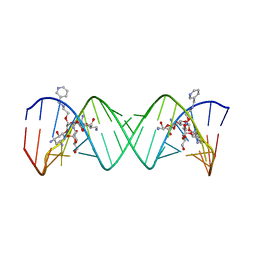 | | Complex Between Paromomycin Derivative JS5-39 and the 16S-Rrna A-Site. | | 分子名称: | (2S,3S,4R,5R,6R)-5-AMINO-2-(AMINOMETHYL)-6-((2R,3R,4R,5S)-5-((1R,2R,3S,5R,6S)-3,5-DIAMINO-2-((2S,3R,4R,5S,6R)-3-AMINO-4 ,5-DIHYDROXY-6-(HYDROXYMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-6-HYDROXYCYCLOHEXYLOXY)-2-(HYDROXYMETHYL)-4-(2-((R)-PIPERIDI N-3-YLMETHYLAMINO)ETHOXY)-TETRAHYDROFURAN-3-YLOXY)-TETRAHYDRO-2H-PYRAN-3,4-DIOL, 5'-R(*CP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3' | | 著者 | Francois, B, Westhof, E. | | 登録日 | 2005-10-21 | | 公開日 | 2005-12-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | Antibacterial aminoglycosides with a modified mode of binding to the ribosomal-RNA decoding site
ANGEW.CHEM.INT.ED.ENGL., 43, 2004
|
|
2BEE
 
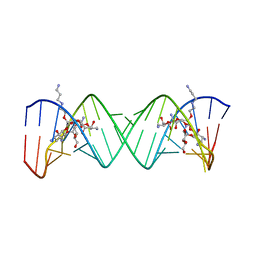 | | Complex Between Paromomycin derivative JS4 and the 16S-Rrna A Site | | 分子名称: | (2S,3S,4R,5R,6R)-5-AMINO-2-(AMINOMETHYL)-6-((2R,3R,4R,5S)-4-(2-(3-AMINOPROPYLAMINO)ETHOXY)-5-((1R,2R,3S,5R,6S)-3,5-DIAM INO-2-((2S,3R,4R,5S,6R)-3-AMINO-4,5-DIHYDROXY-6-(HYDROXYMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-6-HYDROXYCYCLOHEXYLOXY)-2-( HYDROXYMETHYL)-TETRAHYDROFURAN-3-YLOXY)-TETRAHYDRO-2H-PYRAN-3,4-DIOL, 5'-R(*CP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3' | | 著者 | Francois, B, Westhof, E. | | 登録日 | 2005-10-24 | | 公開日 | 2005-12-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Antibacterial aminoglycosides with a modified mode of binding to the ribosomal-RNA decoding site
ANGEW.CHEM.INT.ED.ENGL., 43, 2004
|
|
5QCS
 
 | | Crystal structure of BACE complex with BMC024 | | 分子名称: | (2R,4S)-N-BUTYL-4-HYDROXY-2-METHYL- 4-((E)-(4AS,12R,15S,17AS)-15-METHYL -14,17-DIOXO-2,3,4,4A,6,9,11,12,13, 14,15,16,17,17A-TETRADECAHYDRO-1H-5 ,10-DITHIA-1,13,16-TRIAZA-BENZOCYCL OPENTADECEN-12-YL)-BUTYRAMIDE, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QD4
 
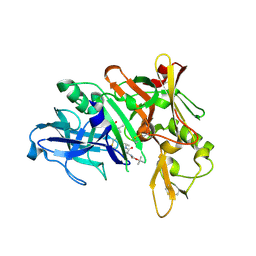 | | Crystal structure of BACE complex with BMC023 | | 分子名称: | Beta-secretase 1, {(E)-(3R,6S,9R)-3-[(1S,3R)-3-((S)-1 -BUTYLCARBAMOYL-2-METHYL-PROPYLCARB AMOYL)-1-HYDROXY-BUTYL]-6-METHYL-5, 8-DIOXO-1,11-DITHIA-4,7-DIAZA-CYCLO PENTADEC-13-EN-9-YL}-CARBAMIC ACID TERT-BUTYL ESTER | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.112 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
