3QMF
 
 | | Crystal strucuture of an inositol monophosphatase family protein (SAS2203) from Staphylococcus aureus MSSA476 | | 分子名称: | Inositol monophosphatase family protein, SULFATE ION | | 著者 | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | 登録日 | 2011-02-04 | | 公開日 | 2012-01-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of Staphylococcal dual specific inositol monophosphatase/NADP(H) phosphatase (SAS2203) delineates the molecular basis of substrate specificity
Biochimie, 94, 2012
|
|
3KSZ
 
 | | Crystal Structure of C151S+H178N mutant of Glyceraldehyde-3-phosphate-dehydrogenase 1 (GAPDH 1) from Staphylococcus aureus MRSA252 complexed with NAD and G3P | | 分子名称: | 3-PHOSPHOGLYCERIC ACID, Glyceraldehyde-3-phosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | 登録日 | 2009-11-24 | | 公開日 | 2010-08-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
3L4S
 
 | | Crystal structure of C151G mutant of Glyceraldehyde 3-phosphate dehydrogenase 1 (GAPDH1) from methicillin resistant Staphylococcus aureus MRSA252 complexed with NAD and G3P | | 分子名称: | 3-PHOSPHOGLYCERIC ACID, Glyceraldehyde-3-phosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | 登録日 | 2009-12-21 | | 公開日 | 2010-08-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
3KSD
 
 | | Crystal Structure of C151S+H178N mutant of Glyceraldehyde-3-phosphate dehydrogenase 1 (GAPDH1) from Staphylococcus aureus MRSA252 complexed with NAD at 2.2 angstrom resolution | | 分子名称: | Glyceraldehyde-3-phosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | 登録日 | 2009-11-22 | | 公開日 | 2010-08-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
3KV3
 
 | | Crystal structure of C151S mutant of Glyceraldehyde-3-phosphate dehydrogenase 1 (GAPDH 1)from methicillin resistant Staphylococcus aureus MRSA252 complexed with NAD and G3P | | 分子名称: | 3-PHOSPHOGLYCERIC ACID, GAPDH, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | 登録日 | 2009-11-29 | | 公開日 | 2010-08-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
3LC7
 
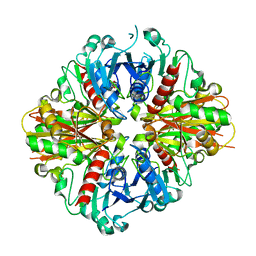 | | Crystal Structure of apo Glyceraldehyde-3-phosphate dehydrogenase 1 (GAPDH1) from methicllin resistant Staphylococcus aureus (MRSA252) | | 分子名称: | GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase 1 | | 著者 | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | 登録日 | 2010-01-10 | | 公開日 | 2010-08-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
3LC2
 
 | | Crystal Structure of Thioacyl-Glyceraldehyde-3-phosphate dehydrogenase 1(GAPDH 1) from methicillin resistant Staphylococcus aureus MRSA252 | | 分子名称: | CHLORIDE ION, GLYCERALDEHYDE-3-PHOSPHATE, GLYCEROL, ... | | 著者 | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | 登録日 | 2010-01-09 | | 公開日 | 2010-08-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
3LC1
 
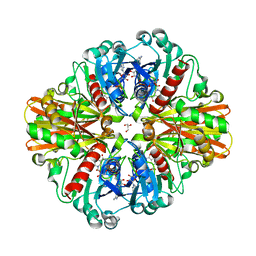 | | Crystal Structure of H178N mutant of Glyceraldehyde-3-phosphate-dehydrogenase 1 (GAPDH 1) from Staphylococcus aureus MRSA252 complexed with NAD at 2.0 angstrom resolution. | | 分子名称: | GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | 登録日 | 2010-01-09 | | 公開日 | 2010-08-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
3L6O
 
 | | Crystal Structure of Phosphate bound apo Glyceraldehyde-3-phosphate dehydrogenase 1 from MRSA252 at 2.2 Angstrom resolution | | 分子名称: | Glyceraldehyde-3-phosphate dehydrogenase 1, PHOSPHATE ION | | 著者 | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | 登録日 | 2009-12-23 | | 公開日 | 2010-08-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
3LVF
 
 | |
3K73
 
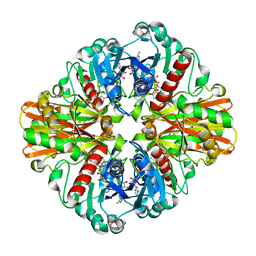 | | Crystal Structure of Phosphate bound Holo Glyceraldehyde-3-phosphate dehydrogenase 1 from MRSA252 at 2.5 Angstrom resolution | | 分子名称: | Glyceraldehyde-3-phosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | 著者 | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | 登録日 | 2009-10-12 | | 公開日 | 2010-08-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
3M9Y
 
 | | Crystal structure of Triosephosphate isomerase from methicillin resistant Staphylococcus aureus at 1.9 Angstrom resolution | | 分子名称: | CITRIC ACID, SODIUM ION, Triosephosphate isomerase | | 著者 | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | 登録日 | 2010-03-23 | | 公開日 | 2011-04-06 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structures of triosephosphate isomerase from methicillin resistant Staphylococcus aureus MRSA252 provide structural insights into novel modes of ligand binding and unique conformations of catalytic loop
Biochimie, 94, 2012
|
|
4DG5
 
 | |
3UWZ
 
 | | Crystal structure of Staphylococcus aureus triosephosphate isomerase complexed with glycerol-2-phosphate | | 分子名称: | 2-HYDROXY-1-(HYDROXYMETHYL)ETHYL DIHYDROGEN PHOSPHATE, PHOSPHATE ION, Triosephosphate isomerase | | 著者 | Mukherjee, S, Roychowdhury, A, Dutta, D, Das, A.K. | | 登録日 | 2011-12-03 | | 公開日 | 2012-10-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structures of triosephosphate isomerase from methicillin resistant Staphylococcus aureus MRSA252 provide structural insights into novel modes of ligand binding and unique conformations of catalytic loop
Biochimie, 94, 2012
|
|
3RYD
 
 | | Crystal structure of Ca bound IMPase family protein from Staphylococcus aureus | | 分子名称: | CALCIUM ION, Inositol monophosphatase family protein, POTASSIUM ION, ... | | 著者 | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | 登録日 | 2011-05-11 | | 公開日 | 2012-01-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Crystal structure of Staphylococcal dual specific inositol monophosphatase/NADP(H) phosphatase (SAS2203) delineates the molecular basis of substrate specificity
Biochimie, 94, 2012
|
|
4OOB
 
 | |
3UWY
 
 | |
3UWU
 
 | | Crystal structure of Staphylococcus Aureus triosephosphate isomerase complexed with glycerol-3-phosphate | | 分子名称: | CITRIC ACID, SN-GLYCEROL-3-PHOSPHATE, Triosephosphate isomerase | | 著者 | Mukherjee, S, Roychowdhury, A, Dutta, D, Das, A.K. | | 登録日 | 2011-12-03 | | 公開日 | 2012-10-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structures of triosephosphate isomerase from methicillin resistant Staphylococcus aureus MRSA252 provide structural insights into novel modes of ligand binding and unique conformations of catalytic loop
Biochimie, 94, 2012
|
|
3VAZ
 
 | | Crystal structure of Staphylococcal GAPDH1 in a hexagonal space group | | 分子名称: | (2R)-2,3-DIHYDROXYPROPANOIC ACID, Glyceraldehyde-3-phosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Roychowdhury, A, Mukherjee, S, Dutta, D, Das, A.K. | | 登録日 | 2011-12-30 | | 公開日 | 2013-01-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.19 Å) | | 主引用文献 | Crystal structure of Staphylococcal GAPDH1 in a hexagonal space group
To be Published
|
|
3UWW
 
 | | Crystal structure of Staphylococcus Aureus triosephosphate isomerase complexed with 3-phosphoglyceric acid | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3-PHOSPHOGLYCERIC ACID, SODIUM ION, ... | | 著者 | Mukherjee, S, Roychowdhury, A, Dutta, D, Das, A.K. | | 登録日 | 2011-12-03 | | 公開日 | 2012-10-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal structures of triosephosphate isomerase from methicillin resistant Staphylococcus aureus MRSA252 provide structural insights into novel modes of ligand binding and unique conformations of catalytic loop
Biochimie, 94, 2012
|
|
3UWV
 
 | | Crystal structure of Staphylococcus Aureus triosephosphate isomerase complexed with 2-phosphoglyceric acid | | 分子名称: | 2-PHOSPHOGLYCERIC ACID, SODIUM ION, Triosephosphate isomerase | | 著者 | Mukherjee, S, Roychowdhury, A, Dutta, D, Das, A.K. | | 登録日 | 2011-12-03 | | 公開日 | 2012-10-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Crystal structures of triosephosphate isomerase from methicillin resistant Staphylococcus aureus MRSA252 provide structural insights into novel modes of ligand binding and unique conformations of catalytic loop
Biochimie, 94, 2012
|
|
3WEW
 
 | |
4ILU
 
 | | Crystal structure of Mycobacterium tuberculosis CarD | | 分子名称: | RNA polymerase-binding transcription factor CarD, SULFATE ION | | 著者 | Thakur, K.G, Kaur, G. | | 登録日 | 2013-01-01 | | 公開日 | 2013-10-30 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of Mycobacterium tuberculosis CarD, an essential RNA polymerase binding protein, reveals a quasidomain-swapped dimeric structural architecture.
Proteins, 82, 2014
|
|
6VUH
 
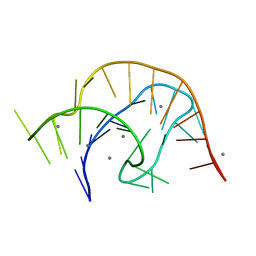 | | APO PreQ1 riboswitch aptamer grown in Mn2+ | | 分子名称: | MANGANESE (II) ION, PREQ1 RIBOSWITCH | | 著者 | Jenkins, J.L, Wedekind, J.E. | | 登録日 | 2020-02-15 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.999 Å) | | 主引用文献 | Analysis of a preQ1-I riboswitch in effector-free and bound states reveals a metabolite-programmed nucleobase-stacking spine that controls gene regulation.
Nucleic Acids Res., 48, 2020
|
|
6VUI
 
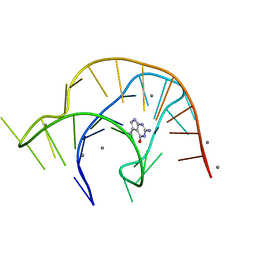 | | Metabolite-bound PreQ1 riboswitch with Mn2+ | | 分子名称: | 7-DEAZA-7-AMINOMETHYL-GUANINE, MANGANESE (II) ION, PREQ1 RIBOSWITCH | | 著者 | Jenkins, J.L, Wedekind, J.E. | | 登録日 | 2020-02-15 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.681 Å) | | 主引用文献 | Analysis of a preQ1-I riboswitch in effector-free and bound states reveals a metabolite-programmed nucleobase-stacking spine that controls gene regulation.
Nucleic Acids Res., 48, 2020
|
|
