5RGD
 
 | | Crystal Structure of Kemp Eliminase HG3.14 with bound transition state analogue, 277K | | 分子名称: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3.14, SULFATE ION | | 著者 | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RG4
 
 | | Crystal Structure of Kemp Eliminase HG3 in unbound state, 277K | | 分子名称: | ACETATE ION, Kemp Eliminase HG3, SULFATE ION | | 著者 | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2021-05-12 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RGC
 
 | | Crystal Structure of Kemp Eliminase HG3.7 with bound transition state analogue, 277K | | 分子名称: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3, SULFATE ION | | 著者 | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2021-05-12 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RGA
 
 | | Crystal Structure of Kemp Eliminase HG3 with bound transition state analogue, 277K | | 分子名称: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3, SULFATE ION | | 著者 | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2021-05-12 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RG7
 
 | | Crystal Structure of Kemp Eliminase HG3.14 in unbound state, 277K | | 分子名称: | Kemp Eliminase HG3, SULFATE ION | | 著者 | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RGE
 
 | | Crystal Structure of Kemp Eliminase HG3.17 with bound transition state analog, 277K | | 分子名称: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3 | | 著者 | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RG9
 
 | | Crystal Structure of Kemp Eliminase HG4 in unbound state, 277K | | 分子名称: | ACETATE ION, Kemp Eliminase HG4, SULFATE ION | | 著者 | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2021-05-12 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RGF
 
 | | Crystal Structure of Kemp Eliminase HG4 with bound transition state analogue, 277K | | 分子名称: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3, SULFATE ION | | 著者 | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
3NF0
 
 | | mPlum-TTN | | 分子名称: | D-MALATE, Fluorescent protein plum | | 著者 | Mayo, S.L, Chica, R.A, Moore, M.M. | | 登録日 | 2010-06-09 | | 公開日 | 2010-10-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural Basis for Changes in the Fluorescent Properties of Computationally Designed Red Fluorescent Proteins
To be Published
|
|
6NJF
 
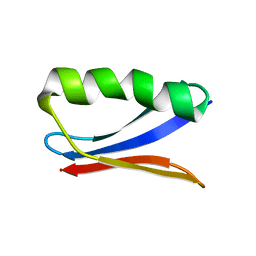 | | Solution NMR Structure of DANCER3-F34A, a rigid and natively folded single mutant of the dynamic protein DANCER-3 | | 分子名称: | Immunoglobulin G-binding protein G | | 著者 | Damry, A.M, Mayer, M.M, Goto, N.K, Chica, R.A. | | 登録日 | 2019-01-03 | | 公開日 | 2019-08-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Origin of conformational dynamics in a globular protein.
Commun Biol, 2, 2019
|
|
5H88
 
 | | Crystal structure of mRojoA mutant - T16V -P63F - W143A - L163V | | 分子名称: | mRojoA fluorescent protein | | 著者 | Pandelieva, A.T, Tremblay, V, Sarvan, S, Chica, R.A, Couture, J.-F. | | 登録日 | 2015-12-23 | | 公開日 | 2016-01-27 | | 最終更新日 | 2016-03-02 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Brighter Red Fluorescent Proteins by Rational Design of Triple-Decker Motif.
Acs Chem.Biol., 11, 2016
|
|
5H89
 
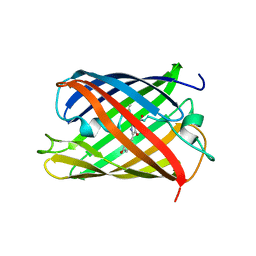 | | Crystal structure of mRojoA mutant - T16V - P63Y - W143G - L163V | | 分子名称: | mRojoA fluorescent protein | | 著者 | Pandelieva, A.T, Tremblay, V, Sarvan, S, Chica, R.A, Couture, J.-F. | | 登録日 | 2015-12-23 | | 公開日 | 2016-01-27 | | 最終更新日 | 2016-03-02 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Brighter Red Fluorescent Proteins by Rational Design of Triple-Decker Motif.
Acs Chem.Biol., 11, 2016
|
|
5H87
 
 | | Crystal structure of mRojoA mutant - P63H - W143S | | 分子名称: | mRojoA fluorescent protein | | 著者 | Pandelieva, A.T, Tremblay, V, Sarvan, S, Chica, R.A, Couture, J.-F. | | 登録日 | 2015-12-23 | | 公開日 | 2016-01-27 | | 最終更新日 | 2016-03-02 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Brighter Red Fluorescent Proteins by Rational Design of Triple-Decker Motif.
Acs Chem.Biol., 11, 2016
|
|
8FOR
 
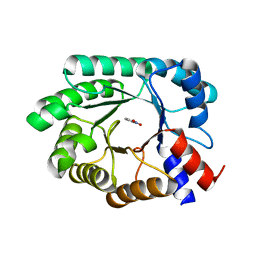 | |
8FMC
 
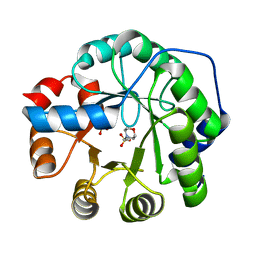 | |
8FMD
 
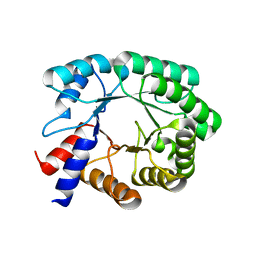 | |
8FOQ
 
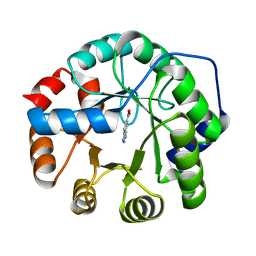 | |
8FME
 
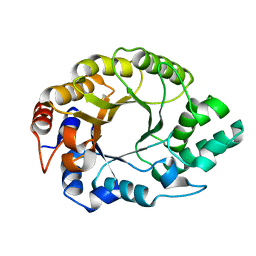 | |
8FOS
 
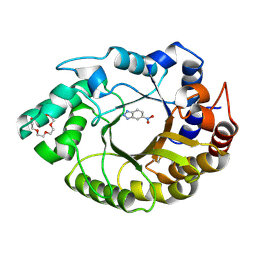 | |
5UBS
 
 | | Solution NMR Structure of NERD-S, a natively folded pentamutant of the B1 domain of streptococcal protein G (GB1) with a solvent-exposed Trp43 | | 分子名称: | Immunoglobulin G-binding protein G | | 著者 | Damry, A.M, Davey, J.A, Goto, N.K, Chica, R.A. | | 登録日 | 2016-12-21 | | 公開日 | 2017-08-23 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Rational design of proteins that exchange on functional timescales.
Nat. Chem. Biol., 13, 2017
|
|
5UCE
 
 | | Solution NMR structure of the major species of DANCER-2, a dynamic and natively folded pentamutant of the B1 domain of streptococcal protein G (GB1) | | 分子名称: | Immunoglobulin G-binding protein G | | 著者 | Damry, A.M, Davey, J.A, Goto, N.K, Chica, R.A. | | 登録日 | 2016-12-22 | | 公開日 | 2017-08-23 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Rational design of proteins that exchange on functional timescales.
Nat. Chem. Biol., 13, 2017
|
|
5UCF
 
 | | Solution NMR-derived model of the minor species of DANCER-2, a dynamic and natively folded pentamutant of the B1 domain of streptococcal protein G (GB1) | | 分子名称: | Immunoglobulin G-binding protein G | | 著者 | Damry, A.M, Davey, J.A, Goto, N.K, Chica, R.A. | | 登録日 | 2016-12-22 | | 公開日 | 2017-08-23 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Rational design of proteins that exchange on functional timescales.
Nat. Chem. Biol., 13, 2017
|
|
5UB0
 
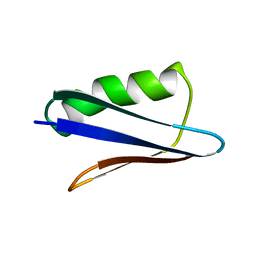 | | Solution NMR Structure of NERD-C, a natively folded tetramutant of the B1 domain of streptococcal protein G (GB1) | | 分子名称: | Immunoglobulin G-binding protein G | | 著者 | Damry, A.M, Davey, J.A, Goto, N.K, Chica, R.A. | | 登録日 | 2016-12-20 | | 公開日 | 2017-08-23 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Rational design of proteins that exchange on functional timescales.
Nat. Chem. Biol., 13, 2017
|
|
6DVS
 
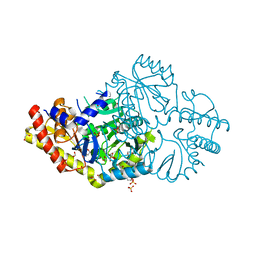 | | Crystal structure of Pseudomonas stutzeri D-phenylglycine aminotransferase | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | 著者 | Couture, J.F, Chica, R. | | 登録日 | 2018-06-25 | | 公開日 | 2018-09-12 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.821 Å) | | 主引用文献 | Structural Determinants of the Stereoinverting Activity of Pseudomonas stutzeri d-Phenylglycine Aminotransferase.
Biochemistry, 57, 2018
|
|
8USK
 
 | |
