4C4W
 
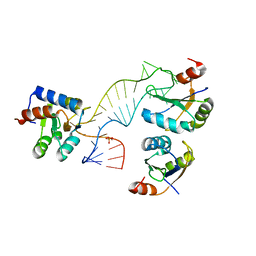 | | Structure of a rare, non-standard sequence k-turn bound by L7Ae protein | | 分子名称: | 50S RIBOSOMAL PROTEIN L7AE, DIHYDROGENPHOSPHATE ION, TSKT-23, ... | | 著者 | Huang, L, Lilley, D.M.J. | | 登録日 | 2013-09-09 | | 公開日 | 2014-02-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Structure of a Rare, Non-Standard Sequence K-Turn Bound by L7Ae Protein
Nucleic Acids Res., 42, 2014
|
|
4B5R
 
 | |
4CS1
 
 | |
4C40
 
 | |
6YL5
 
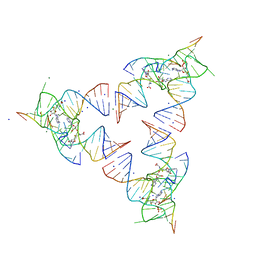 | | Crystal structure of the SAM-SAH riboswitch with SAH | | 分子名称: | Chains: A,B,C,D,E,F,G,H,I,J,K,L, MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | Huang, L, Lilley, D.M.J. | | 登録日 | 2020-04-06 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 2020
|
|
6YMI
 
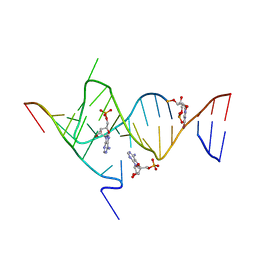 | | Crystal structure of the SAM-SAH riboswitch with AMP. | | 分子名称: | 5-BROMOCYTIDINE 5'-(DIHYDROGEN PHOSPHATE), ADENOSINE MONOPHOSPHATE, Chains: A,C,F,I,M,O, ... | | 著者 | Huang, L, Lilley, D.M.J. | | 登録日 | 2020-04-08 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 48, 2020
|
|
6YMJ
 
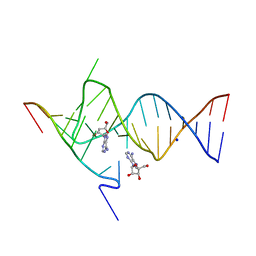 | | Crystal structure of the SAM-SAH riboswitch with adenosine. | | 分子名称: | 5-BROMOCYTIDINE 5'-(DIHYDROGEN PHOSPHATE), ADENOSINE, Chains: A,C,F,I,M,O, ... | | 著者 | Huang, L, Lilley, D.M.J. | | 登録日 | 2020-04-08 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 48, 2020
|
|
6YLB
 
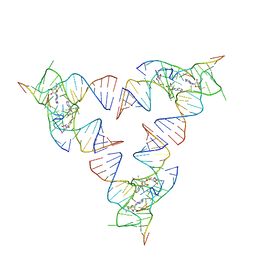 | | Crystal structure of the SAM-SAH riboswitch with SAM | | 分子名称: | Chains: A,C,F,I,M,O, Chains: B,D,G,J,N,P, S-ADENOSYLMETHIONINE | | 著者 | Huang, L, Lilley, D.M.J. | | 登録日 | 2020-04-07 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 2020
|
|
6YML
 
 | |
6YMK
 
 | | Crystal structure of the SAM-SAH riboswitch with AMP | | 分子名称: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, Chains: A,C,F,I,M,O, Chains: B,D,G,J,N,P, ... | | 著者 | Huang, L, Lilley, D.M.J. | | 登録日 | 2020-04-08 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 48, 2020
|
|
6YMM
 
 | |
2YDH
 
 | |
6TF0
 
 | | Crystal structure of the ADP-binding domain of the NAD+ riboswitch with Nicotinamide adenine dinucleotide, reduced (NADH) | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Chains: A, MAGNESIUM ION, ... | | 著者 | Huang, L, Lilley, D.M.J. | | 登録日 | 2019-11-12 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure and ligand binding of the ADP-binding domain of the NAD+ riboswitch.
Rna, 26, 2020
|
|
6TF3
 
 | |
6TFE
 
 | |
6TB7
 
 | |
6TF2
 
 | |
6TFH
 
 | |
6TFG
 
 | |
6TF1
 
 | |
6TFF
 
 | |
5FKE
 
 | |
5FJ1
 
 | |
5FKF
 
 | |
5FK2
 
 | |
