7UU8
 
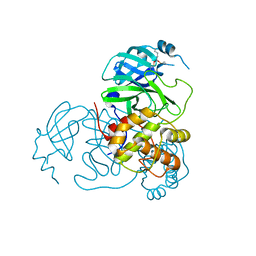 | |
7UUD
 
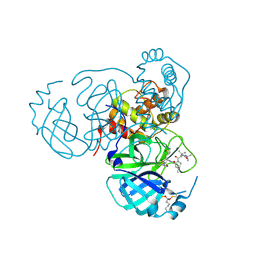 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI33 | | 分子名称: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S,3R)-4-(ethylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Yang, K.S, Liu, W.R. | | 登録日 | 2022-04-28 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | A Novel Y-Shaped, S-O-N-O-S-Bridged Cross-Link between Three Residues C22, C44, and K61 Is Frequently Observed in the SARS-CoV-2 Main Protease.
Acs Chem.Biol., 18, 2023
|
|
7UUC
 
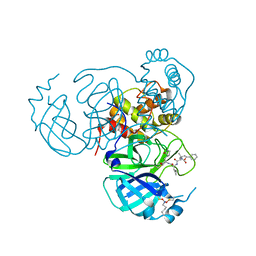 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI19 | | 分子名称: | 3C-like proteinase nsp5, N-[(benzyloxy)carbonyl]-3-methyl-L-valyl-3-cyclopropyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | 著者 | Yang, K.S, Liu, W.R. | | 登録日 | 2022-04-28 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A Novel Y-Shaped, S-O-N-O-S-Bridged Cross-Link between Three Residues C22, C44, and K61 Is Frequently Observed in the SARS-CoV-2 Main Protease.
Acs Chem.Biol., 18, 2023
|
|
7UU7
 
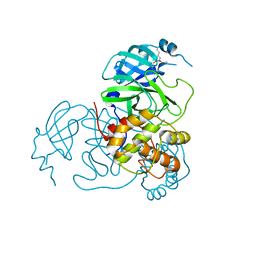 | |
7WL3
 
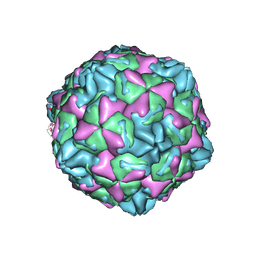 | | CVB5 expended empty particle | | 分子名称: | Capsid protein, Genome polyprotein | | 著者 | Yang, P, Wang, K. | | 登録日 | 2022-01-12 | | 公開日 | 2022-03-30 | | 最終更新日 | 2022-10-12 | | 実験手法 | ELECTRON MICROSCOPY (2.95 Å) | | 主引用文献 | Atomic Structures of Coxsackievirus B5 Provide Key Information on Viral Evolution and Survival.
J.Virol., 96, 2022
|
|
7DO4
 
 | | Crystal structure of CD97-CD55 complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Niu, M, Song, G. | | 登録日 | 2020-12-11 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural basis for CD97 recognition of the decay-accelerating factor CD55 suggests mechanosensitive activation of adhesion GPCRs.
J.Biol.Chem., 296, 2021
|
|
7EAK
 
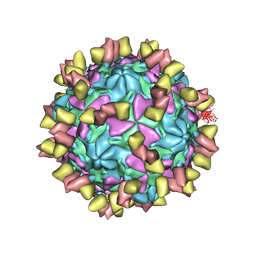 | | Echovirus3 full particle in complex with 5G3 fab | | 分子名称: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | 著者 | Feng, R. | | 登録日 | 2021-03-07 | | 公開日 | 2021-07-14 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural basis for neutralization of an anicteric hepatitis associated echovirus by a potent neutralizing antibody.
Cell Discov, 7, 2021
|
|
7EAH
 
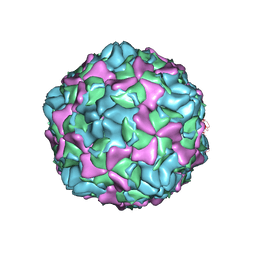 | | Echovirus3 empty expanded particle | | 分子名称: | Capsid protein VP0, Capsid protein VP1, Capsid protein VP3 | | 著者 | Feng, R. | | 登録日 | 2021-03-07 | | 公開日 | 2021-07-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis for neutralization of an anicteric hepatitis associated echovirus by a potent neutralizing antibody.
Cell Discov, 7, 2021
|
|
7EAI
 
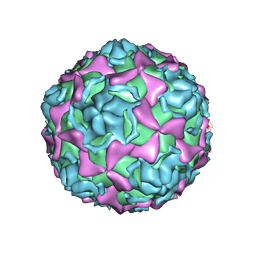 | | Echovirus3 empty compacted particle | | 分子名称: | Capsid protein VP0, Capsid protein VP1, Capsid protein VP3, ... | | 著者 | Feng, R. | | 登録日 | 2021-03-07 | | 公開日 | 2021-07-14 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis for neutralization of an anicteric hepatitis associated echovirus by a potent neutralizing antibody.
Cell Discov, 7, 2021
|
|
7EAJ
 
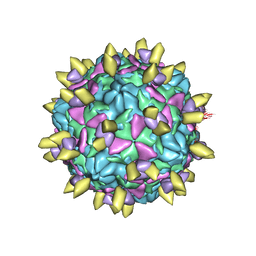 | |
5YPS
 
 | | The structural basis of histone chaperoneVps75 | | 分子名称: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Chen, Y, Zhang, Y, Dou, Y, Wang, M, Xu, S, Jiang, H, Limper, A, Su, D. | | 登録日 | 2017-11-03 | | 公開日 | 2018-11-07 | | 最終更新日 | 2020-06-10 | | 実験手法 | X-RAY DIFFRACTION (2.097 Å) | | 主引用文献 | Structural basis for the acetylation of histone H3K9 and H3K27 mediated by the histone chaperone Vps75 inPneumocystis carinii.
Signal Transduct Target Ther, 4, 2019
|
|
5ZB5
 
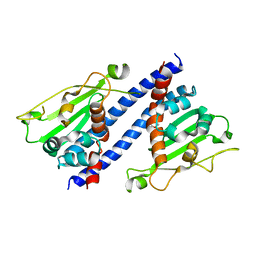 | | The structural basis of histone chaperoneVps75 | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, NAP family histone chaperone vps75 | | 著者 | Chen, Y, Zhang, Y, Dou, Y, Wang, M, Xu, S, Jiang, H, Limper, A, Su, D. | | 登録日 | 2018-02-09 | | 公開日 | 2019-02-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.299 Å) | | 主引用文献 | Structural basis for the acetylation of histone H3K9 and H3K27 mediated by the histone chaperone Vps75 inPneumocystis carinii.
Signal Transduct Target Ther, 4, 2019
|
|
