4ZRO
 
 | | 2.1 A X-Ray Structure of FIPV-3CLpro bound to covalent inhibitor | | 分子名称: | 3C-like proteinase, Bounded inhibitor of N-(tert-butoxycarbonyl)-L-seryl-L-valyl-N-{(2S)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-leucinamide, DIMETHYL SULFOXIDE | | 著者 | St John, S.E, Mesecar, A.D. | | 登録日 | 2015-05-12 | | 公開日 | 2015-10-14 | | 最終更新日 | 2019-12-11 | | 実験手法 | X-RAY DIFFRACTION (2.0566 Å) | | 主引用文献 | X-ray structure and inhibition of the feline infectious peritonitis virus 3C-like protease: Structural implications for drug design.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4WF1
 
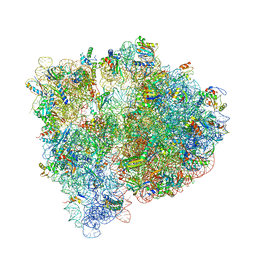 | | Crystal structure of the E. coli ribosome bound to negamycin. | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Olivier, N.B, Altman, R.B, Noeske, J, Basarab, G.S, Code, E, Ferguson, A.D, Gao, N, Huang, J, Juette, M.F, Livchak, S, Miller, M.D, Prince, D.B, Cate, J.H.D, Buurman, E.T, Blanchard, S.C. | | 登録日 | 2014-09-11 | | 公開日 | 2014-11-05 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.09 Å) | | 主引用文献 | Negamycin induces translational stalling and miscoding by binding to the small subunit head domain of the Escherichia coli ribosome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MAZ
 
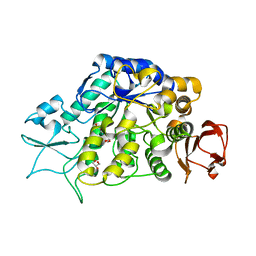 | | The Structure of MalL mutant enzyme V200S from Bacillus subtilus | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Hobbs, J.K, Jiao, W, Easter, A.D, Parker, E.J, Schipper, L.A, Arcus, V.L. | | 登録日 | 2013-08-18 | | 公開日 | 2013-09-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Change in heat capacity for enzyme catalysis determines temperature dependence of enzyme catalyzed rates.
Acs Chem.Biol., 8, 2013
|
|
4MB1
 
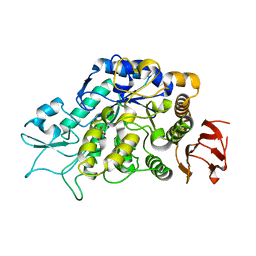 | | The Structure of MalL mutant enzyme G202P from Bacillus subtilus | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Oligo-1,6-glucosidase 1 | | 著者 | Hobbs, J.K, Jiao, W, Easter, A.D, Parker, E.J, Schipper, L.A, Arcus, V.L. | | 登録日 | 2013-08-19 | | 公開日 | 2013-09-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Change in heat capacity for enzyme catalysis determines temperature dependence of enzyme catalyzed rates.
Acs Chem.Biol., 8, 2013
|
|
4M8B
 
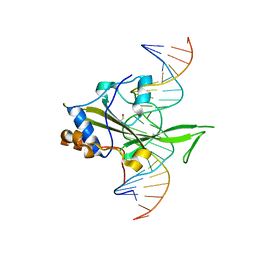 | | Fungal Protein | | 分子名称: | 1,2-ETHANEDIOL, Chain A of dsDNA containing the cis-regulatory element, Chain B of dsDNA containing the cis-regulatory element, ... | | 著者 | Rosenberg, O.S, Lohse, M.L, Stroud, R.M, Johnson, A.D. | | 登録日 | 2013-08-13 | | 公開日 | 2014-06-25 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Structure of a new DNA-binding domain which regulates pathogenesis in a wide variety of fungi.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4WZ4
 
 | | Crystal structure of P. aeruginosa AmpC | | 分子名称: | Beta-lactamase, GLYCEROL, {(3R)-6-[(3-amino-1,2,4-thiadiazol-5-yl)oxy]-1-hydroxy-4,5-dimethyl-1,3-dihydro-2,1-benzoxaborol-3-yl}acetic acid | | 著者 | Ferguson, A.D. | | 登録日 | 2014-11-18 | | 公開日 | 2015-08-05 | | 最終更新日 | 2018-04-25 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | 4,5-Disubstituted 6-Aryloxy-1,3-dihydrobenzo[c][1,2]oxaboroles Are Broad-Spectrum Serine beta-Lactamase Inhibitors.
ACS Infect Dis, 1, 2015
|
|
4M56
 
 | | The Structure of Wild-type MalL from Bacillus subtilis | | 分子名称: | D-glucose, GLYCEROL, Oligo-1,6-glucosidase 1, ... | | 著者 | Hobbs, J.K, Jiao, W, Easter, A.D, Parker, E.J, Schipper, L.A, Arcus, V.L. | | 登録日 | 2013-08-08 | | 公開日 | 2013-10-02 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Change in heat capacity for enzyme catalysis determines temperature dependence of enzyme catalyzed rates.
Acs Chem.Biol., 8, 2013
|
|
4M6V
 
 | |
4X7D
 
 | |
4MFD
 
 | |
4LOC
 
 | |
4WZ5
 
 | | Crystal structure of P. aeruginosa OXA10 | | 分子名称: | Beta-lactamase OXA-10, CARBON DIOXIDE, SULFATE ION, ... | | 著者 | Ferguson, A.D. | | 登録日 | 2014-11-18 | | 公開日 | 2015-08-05 | | 最終更新日 | 2016-09-28 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | 4,5-Disubstituted 6-Aryloxy-1,3-dihydrobenzo[c][1,2]oxaboroles Are Broad-Spectrum Serine beta-Lactamase Inhibitors.
Acs Infect Dis., 1, 2015
|
|
4X7C
 
 | |
4X7E
 
 | |
5BSX
 
 | |
4Z4W
 
 | | Crystal structure of GII.10 P domain in complex with 4.7mM fucose | | 分子名称: | 1,2-ETHANEDIOL, Capsid protein, beta-L-fucopyranose | | 著者 | Koromyslova, A.D, Leuthold, M.M, Hansman, G.S. | | 登録日 | 2015-04-02 | | 公開日 | 2015-05-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The sweet quartet: Binding of fucose to the norovirus capsid.
Virology, 483, 2015
|
|
4WYY
 
 | | Crystal Structure of P. aeruginosa AmpC | | 分子名称: | Beta-lactamase, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Ferguson, A.D. | | 登録日 | 2014-11-18 | | 公開日 | 2015-08-05 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | 4,5-Disubstituted 6-Aryloxy-1,3-dihydrobenzo[c][1,2]oxaboroles Are Broad-Spectrum Serine beta-Lactamase Inhibitors.
ACS Infect Dis, 1, 2015
|
|
5AKB
 
 | | MutS in complex with the N-terminal domain of MutL - crystal form 1 | | 分子名称: | DNA MISMATCH REPAIR PROTEIN MUTL, DNA MISMATCH REPAIR PROTEIN MUTS, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Groothuizen, F.S, Winkler, I, Cristovao, M, Fish, A, Winterwerp, H.H.K, Reumer, A, Marx, A.D, Hermans, N, Nicholls, R.A, Murshudov, G.N, Lebbink, J.H.G, Friedhoff, P, Sixma, T.K. | | 登録日 | 2015-03-03 | | 公開日 | 2015-07-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (4.71 Å) | | 主引用文献 | MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA.
Elife, 4, 2015
|
|
6SCT
 
 | | Cryo-EM structure of the consensus triskelion hub of the clathrin coat complex | | 分子名称: | Clathrin heavy chain, Clathrin light chain | | 著者 | Morris, K.L, Cameron, A.D, Sessions, R, Smith, C.J. | | 登録日 | 2019-07-25 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.69 Å) | | 主引用文献 | Cryo-EM of multiple cage architectures reveals a universal mode of clathrin self-assembly.
Nat.Struct.Mol.Biol., 26, 2019
|
|
5AKC
 
 | | MutS in complex with the N-terminal domain of MutL - crystal form 2 | | 分子名称: | DNA MISMATCH REPAIR PROTEIN MUTL, DNA MISMATCH REPAIR PROTEIN MUTS, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Groothuizen, F.S, Winkler, I, Cristovao, M, Fish, A, Winterwerp, H.H.K, Reumer, A, Marx, A.D, Hermans, N, Nicholls, R.A, Murshudov, G.N, Lebbink, J.H.G, Friedhoff, P, Sixma, T.K. | | 登録日 | 2015-03-03 | | 公開日 | 2015-07-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (6.6 Å) | | 主引用文献 | MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA.
Elife, 4, 2015
|
|
5AKD
 
 | | MutS in complex with the N-terminal domain of MutL - crystal form 3 | | 分子名称: | DNA MISMATCH REPAIR PROTEIN MUTL, DNA MISMATCH REPAIR PROTEIN MUTS, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Groothuizen, F.S, Winkler, I, Cristovao, M, Fish, A, Winterwerp, H.H.K, Reumer, A, Marx, A.D, Hermans, N, Nicholls, R.A, Murshudov, G.N, Lebbink, J.H.G, Friedhoff, P, Sixma, T.K. | | 登録日 | 2015-03-03 | | 公開日 | 2015-07-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (7.6 Å) | | 主引用文献 | MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA.
Elife, 4, 2015
|
|
4MFE
 
 | |
4MIM
 
 | |
4Z4S
 
 | | Crystal structure of GII.10 P domain in complex with 150mM fucose | | 分子名称: | 1,2-ETHANEDIOL, Capsid protein, NITRATE ION, ... | | 著者 | Koromyslova, A.D, Leuthold, M.M, Hansman, G.S. | | 登録日 | 2015-04-02 | | 公開日 | 2015-05-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The sweet quartet: Binding of fucose to the norovirus capsid.
Virology, 483, 2015
|
|
4MKJ
 
 | | Crystal structure of L-methionine gamma-lyase from Citrobacter freundii modified by allicine | | 分子名称: | Methionine gamma-lyase, PENTAETHYLENE GLYCOL, SODIUM ION, ... | | 著者 | Revtovich, S.V, Nikulin, A.D, Morozova, E.A, Zakomirdina, L.N, Demidkina, T.V. | | 登録日 | 2013-09-05 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.849 Å) | | 主引用文献 | Alliin is a suicide substrate of Citrobacter freundii methionine gamma-lyase: structural bases of inactivation of the enzyme.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
