8JCK
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with Compound 32 | | 分子名称: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Zhao, Y, Zhu, Y, Rao, Z. | | 登録日 | 2023-05-11 | | 公開日 | 2024-05-15 | | 最終更新日 | 2025-05-28 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | De novo design of SARS-CoV-2 main protease inhibitors with characteristic binding modes.
Structure, 32, 2024
|
|
8JCM
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with Compound 55 | | 分子名称: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, HYDROSULFURIC ACID, ... | | 著者 | Zhao, Y, Zhu, Y, Rao, Z. | | 登録日 | 2023-05-11 | | 公開日 | 2024-05-15 | | 最終更新日 | 2025-05-28 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | De novo design of SARS-CoV-2 main protease inhibitors with characteristic binding modes.
Structure, 32, 2024
|
|
8JCO
 
 | |
8JCL
 
 | |
8JCJ
 
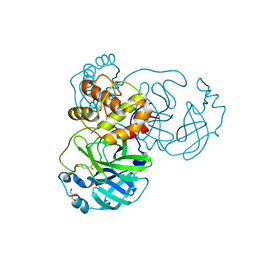 | | The crystal structure of SARS-CoV-2 main protease in complex with Compound 18 | | 分子名称: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Zhao, Y, Zhu, Y, Rao, Z. | | 登録日 | 2023-05-11 | | 公開日 | 2024-05-15 | | 最終更新日 | 2025-05-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | De novo design of SARS-CoV-2 main protease inhibitors with characteristic binding modes.
Structure, 32, 2024
|
|
6NJM
 
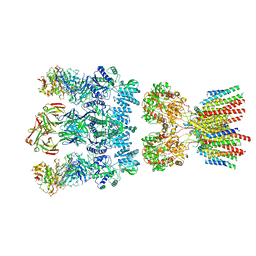 | |
6NJN
 
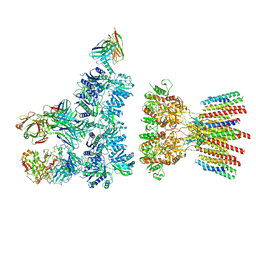 | |
6NJL
 
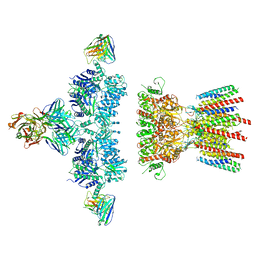 | |
8JGR
 
 | |
8JGT
 
 | |
8JGX
 
 | |
8JGW
 
 | |
8JGU
 
 | |
8JGQ
 
 | |
8JGP
 
 | |
8JGO
 
 | |
8WTY
 
 | |
8WTX
 
 | |
8WTV
 
 | | Cryo-EM structure of noradrenaline transporter in complex with noradrenaline | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Noradrenaline, ... | | 著者 | Zhao, Y, Hu, T, Yu, Z. | | 登録日 | 2023-10-19 | | 公開日 | 2024-08-07 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Transport and inhibition mechanisms of the human noradrenaline transporter.
Nature, 632, 2024
|
|
8XTX
 
 | | structure of a protein | | 分子名称: | Green fluorescent protein,Vesicular acetylcholine transporter,antibody | | 著者 | Zhao, Y, Ma, Q, Dong, Y, Meng, Y. | | 登録日 | 2024-01-12 | | 公開日 | 2024-12-25 | | 最終更新日 | 2025-06-11 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Binding mechanism and antagonism of the vesicular acetylcholine transporter VAChT.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8XTW
 
 | | structure of a protein | | 分子名称: | ACETYLCHOLINE, Green fluorescent protein,Vesicular acetylcholine transporter,antibody | | 著者 | Zhao, Y, Ma, Q, Dong, Y, Meng, Y. | | 登録日 | 2024-01-12 | | 公開日 | 2024-12-25 | | 最終更新日 | 2025-06-11 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Binding mechanism and antagonism of the vesicular acetylcholine transporter VAChT.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8XTY
 
 | | structure of a protein | | 分子名称: | Green fluorescent protein,Vesicular acetylcholine transporter,Green fluorescent protein,Vesicular acetylcholine transporter,Green fluorescent protein,Vesicular acetylcholine transporter,Green fluorescent protein,Vesicular acetylcholine transporter,antibody, vesamicol | | 著者 | Zhao, Y, Ma, Q, Dong, Y, Meng, Y. | | 登録日 | 2024-01-12 | | 公開日 | 2024-12-25 | | 最終更新日 | 2025-06-11 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Binding mechanism and antagonism of the vesicular acetylcholine transporter VAChT.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8Y2C
 
 | |
8Y2E
 
 | |
8Y2G
 
 | |
