3X2L
 
 | | X-ray structure of PcCel45A apo form at 95K. | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-methylpentane-1,5-diol, Endoglucanase V-like protein | | 著者 | Nakamura, A, Ishida, T, Ohta, K, Tanaka, H, Inaka, K, Samejima, M, Igarashi, K. | | 登録日 | 2014-12-22 | | 公開日 | 2015-10-14 | | 最終更新日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (0.83 Å) | | 主引用文献 | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
3X2M
 
 | | X-ray structure of PcCel45A with cellopentaose at 0.64 angstrom resolution. | | 分子名称: | Endoglucanase V-like protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Nakamura, A, Ishida, T, Samejima, M, Igarashi, K. | | 登録日 | 2014-12-22 | | 公開日 | 2015-10-14 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (0.64 Å) | | 主引用文献 | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
3X2K
 
 | | X-ray structure of PcCel45A D114N with cellopentaose at 95K. | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-methylpentane-1,5-diol, Endoglucanase V-like protein, ... | | 著者 | Nakamura, A, Ishida, T, Samejima, M, Igarashi, K. | | 登録日 | 2014-12-22 | | 公開日 | 2015-10-07 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.182 Å) | | 主引用文献 | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
3X2I
 
 | | X-ray structure of PcCel45A N92D apo form at 298K. | | 分子名称: | 3-methylpentane-1,5-diol, Endoglucanase V-like protein | | 著者 | Nakamura, A, Ishida, T, Samejima, M, Igarashi, K. | | 登録日 | 2014-12-22 | | 公開日 | 2015-10-07 | | 最終更新日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
3X2H
 
 | | X-ray structure of PcCel45A N92D with cellopentaose at 95K. | | 分子名称: | 3-methylpentane-1,5-diol, Endoglucanase V-like protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | 著者 | Nakamura, A, Ishida, T, Samejima, M, Igarashi, K. | | 登録日 | 2014-12-22 | | 公開日 | 2015-10-07 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (0.99 Å) | | 主引用文献 | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
3X2J
 
 | | X-ray structure of PcCel45A D114N apo form at 95K. | | 分子名称: | 3-methylpentane-1,5-diol, Endoglucanase V-like protein | | 著者 | Nakamura, A, Ishida, T, Samejima, M, Igarashi, K. | | 登録日 | 2014-12-22 | | 公開日 | 2015-10-07 | | 最終更新日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (1.301 Å) | | 主引用文献 | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
3X2G
 
 | | X-ray structure of PcCel45A N92D apo form at 100K | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-methylpentane-1,5-diol, Endoglucanase V-like protein | | 著者 | Nakamura, A, Ishida, T, Samejima, M, Igarashi, K. | | 登録日 | 2014-12-22 | | 公開日 | 2015-10-07 | | 最終更新日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
3X2N
 
 | | Proton relay pathway in inverting cellulase | | 分子名称: | Endoglucanase V-like protein, SULFATE ION | | 著者 | Nakamura, A, Ishida, T, Fushinobu, S, Igarashi, K, Samejima, M. | | 登録日 | 2014-12-22 | | 公開日 | 2015-10-14 | | 最終更新日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
5YGY
 
 | | Crystal Structure of BACE1 in complex with (S)-N-(3-(2-amino-6-(fluoromethyl)-4 -methyl-4H-1,3-oxazin-4-yl)-4-fluorophenyl)-5-cyanopicolinamide | | 分子名称: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | 著者 | Fuchino, K, Mitsuoka, Y, Masui, M, Kurose, N, Yoshida, S, Komano, K, Yamamoto, T, Ogawa, M, Unemura, C, Hosono, M, Ito, H, Sakaguchi, G, Ando, S, Ohnishi, S, Kido, Y, Fukushima, T, Miyajima, H, Hiroyama, S, Koyabu, K, Dhuyvetter, D, Borghys, H, Gijsen, H, Yamano, Y, Iso, Y, Kusakabe, K. | | 登録日 | 2017-09-27 | | 公開日 | 2018-05-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Rational Design of Novel 1,3-Oxazine Based beta-Secretase (BACE1) Inhibitors: Incorporation of a Double Bond To Reduce P-gp Efflux Leading to Robust A beta Reduction in the Brain
J. Med. Chem., 61, 2018
|
|
3SAK
 
 | |
8RKI
 
 | | Molecular basis of ZP3/ZP1 heteropolymerization: crystal structure of a native vertebrate egg coat filament fragment | | 分子名称: | Choriogenin H, YTTERBIUM (III) ION, Zona pellucida sperm-binding protein 3, ... | | 著者 | Wiseman, B, Zamora-Caballero, S, de Sanctis, D, Yasumasu, S, Jovine, L. | | 登録日 | 2023-12-25 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (4.2 Å) | | 主引用文献 | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
1C4E
 
 | |
5YGX
 
 | | Structure of BACE1 in complex with N-(3-((4R,5R,6S)-2-amino-6-(1,1-difluoroethyl)-5-fluoro-4-methyl-5,6-dihydro-4H-1,3-oxazin-4-yl)-4-fluorophenyl)-5-(fluoromethoxy)pyrazine-2-carboxamide | | 分子名称: | Beta-secretase 1, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Nakahara, K, Fuchino, K, Komano, K, Asada, N, Tadano, G, Hasegawa, T, Yamamoto, T, Sako, Y, Ogawa, M, Unemura, C, Hosono, M, Sakaguchi, G, Ando, S, Ohnishi, S, Kido, Y, Fukushima, T, Dhuyvetter, D, Borghys, H, Gijsen, H, Yamano, Y, Iso, Y, Kusakabe, K. | | 登録日 | 2017-09-27 | | 公開日 | 2018-08-08 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Discovery of Potent and Centrally Active 6-Substituted 5-Fluoro-1,3-dihydro-oxazine beta-Secretase (BACE1) Inhibitors via Active Conformation Stabilization
J. Med. Chem., 61, 2018
|
|
6JT4
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(4S,6S)-2-amino-4-methyl-6-(trifluoromethyl)-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | 分子名称: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | 著者 | Anan, K, Iso, Y, Oguma, T, Nakahara, K, Suzuki, S, Yamamoto, T, Matsuoka, E, Ito, H, Sakaguchi, G, Ando, S, Morimoto, K, Kanegawa, N, Kido, Y, Kawachi, T, Fukushima, T, Teisman, A, Urmaliya, V, Dhuyvetter, D, Borghys, H, Austin, N, Bergh, A.V.D, Verboven, P, Bischoff, F, Gijsen, H.J.M, Yamano, Y, Kusakabe, K.I. | | 登録日 | 2019-04-08 | | 公開日 | 2020-01-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Trifluoromethyl Dihydrothiazine-Based beta-Secretase (BACE1) Inhibitors with Robust Central beta-Amyloid Reduction and Minimal Covalent Binding Burden.
Chemmedchem, 14, 2019
|
|
1SAL
 
 | |
1SAK
 
 | |
1SAE
 
 | |
1SAF
 
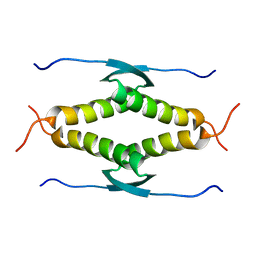 | |
8BQU
 
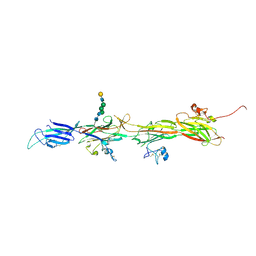 | | Molecular basis of ZP3/ZP1 heteropolymerization: crystal structure of a native vertebrate egg coat filament | | 分子名称: | Choriogenin H, Zona pellucida sperm-binding protein 3, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Bokhove, M, de Sanctis, D, Yasumasu, S, Jovine, L. | | 登録日 | 2022-11-21 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
5X9O
 
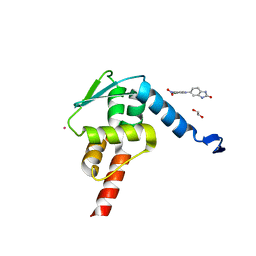 | | Crystal structure of the BCL6 BTB domain in complex with Compound 1a | | 分子名称: | 1,2-ETHANEDIOL, 5-[(2-chloranyl-4-nitro-phenyl)amino]-1,3-dihydrobenzimidazol-2-one, B-cell lymphoma 6 protein, ... | | 著者 | Sogabe, S, Ida, K, Lane, W, Snell, G. | | 登録日 | 2017-03-08 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Discovery of a novel B-cell lymphoma 6 (BCL6)-corepressor interaction inhibitor by utilizing structure-based drug design
Bioorg. Med. Chem., 25, 2017
|
|
5X9P
 
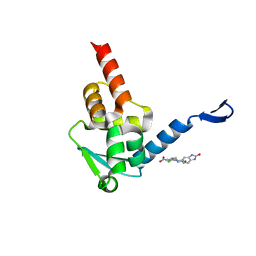 | | Crystal structure of the BCL6 BTB domain in complex with Compound 5 | | 分子名称: | 3-[[4-chloranyl-2-nitro-5-[(2-oxidanylidene-1,3-dihydrobenzimidazol-5-yl)amino]phenyl]amino]propanoic acid, B-cell lymphoma 6 protein | | 著者 | Sogabe, S, Ida, K, Lane, W, Snell, G. | | 登録日 | 2017-03-08 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Discovery of a novel B-cell lymphoma 6 (BCL6)-corepressor interaction inhibitor by utilizing structure-based drug design
Bioorg. Med. Chem., 25, 2017
|
|
5X4N
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 4 | | 分子名称: | 1,2-ETHANEDIOL, 5-chloro-N4-phenylpyrimidine-2,4-diamine, B-cell lymphoma 6 protein | | 著者 | Sogabe, S, Ida, K, Lane, W, Snell, G. | | 登録日 | 2017-02-13 | | 公開日 | 2017-05-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
5X4O
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 5 | | 分子名称: | B-cell lymphoma 6 protein, N-methyl-N-{3-[({2-[(2-oxo-2,3-dihydro-1H-indol-5-yl)amino]-5-(trifluoromethyl)pyrimidin-4-yl}amino)methyl]pyridin-2-yl}methanesulfonamide | | 著者 | Sogabe, S, Ida, K, Lane, W, Snell, G. | | 登録日 | 2017-02-13 | | 公開日 | 2017-05-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
5X4Q
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 7 | | 分子名称: | 1,2-ETHANEDIOL, 5-[[5-chloranyl-2-(pyridin-3-ylmethylamino)pyrimidin-4-yl]amino]-1,3-dihydroindol-2-one, B-cell lymphoma 6 protein, ... | | 著者 | Sogabe, S, Ida, K, Lane, W, Snell, G. | | 登録日 | 2017-02-13 | | 公開日 | 2017-05-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
5X4M
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 1 | | 分子名称: | B-cell lymphoma 6 protein, N-phenyl-1,3,5-triazine-2,4-diamine | | 著者 | Sogabe, S, Ida, K, Lane, W, Snell, G. | | 登録日 | 2017-02-13 | | 公開日 | 2017-05-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
