7XJJ
 
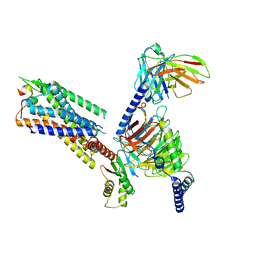 | | Cryo-EM structure of the galanin-bound GALR1-miniGo complex | | Descriptor: | G protein subunit alpha o1,Guanine nucleotide-binding protein G(o) subunit alpha, Galanin, Galanin receptor type 1, ... | | Authors: | Jiang, W, Zheng, S. | | Deposit date: | 2022-04-18 | | Release date: | 2023-05-03 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into galanin receptor signaling.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WQ3
 
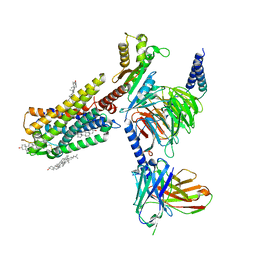 | | Galanin-bound galanin receptor 1 in complex with Gi | | Descriptor: | CHOLESTEROL, Galanin, Galanin receptor type 1, ... | | Authors: | Duan, J, Shen, D.D, Xu, H.E, Zhang, Y, Jiang, Y. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-20 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular basis for allosteric agonism and G protein subtype selectivity of galanin receptors
Nat Commun, 13, 2022
|
|
