1AHN
 
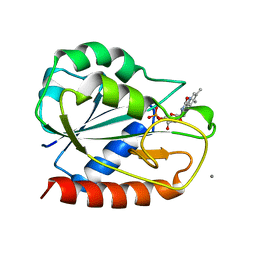 | | E. COLI FLAVODOXIN AT 2.6 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Hoover, D.M, Ludwig, M.L. | | Deposit date: | 1997-04-07 | | Release date: | 1997-12-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A flavodoxin that is required for enzyme activation: the structure of oxidized flavodoxin from Escherichia coli at 1.8 A resolution.
Protein Sci., 6, 1997
|
|
1AHO
 
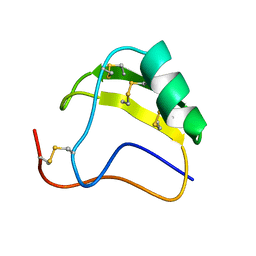 | | THE AB INITIO STRUCTURE DETERMINATION AND REFINEMENT OF A SCORPION PROTEIN TOXIN | | Descriptor: | TOXIN II | | Authors: | Smith, G.D, Blessing, R.H, Ealick, S.E, Fontecilla-Camps, J.C, Hauptman, H.A, Housset, D, Langs, D.A, Miller, R. | | Deposit date: | 1997-04-08 | | Release date: | 1997-10-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Ab initio structure determination and refinement of a scorpion protein toxin.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1AHP
 
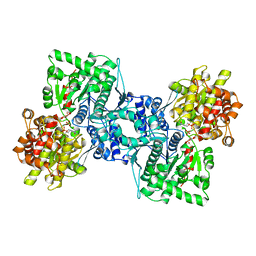 | | OLIGOSACCHARIDE SUBSTRATE BINDING IN ESCHERICHIA COLI MALTODEXTRIN PHSPHORYLASE | | Descriptor: | E.COLI MALTODEXTRIN PHOSPHORYLASE, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | O'Reilly, M, Watson, K.A, Schinzel, R, Palm, D, Johnson, L.N. | | Deposit date: | 1997-04-10 | | Release date: | 1997-10-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Oligosaccharide substrate binding in Escherichia coli maltodextrin phosphorylase.
Nat.Struct.Biol., 4, 1997
|
|
1AHS
 
 | |
1AI1
 
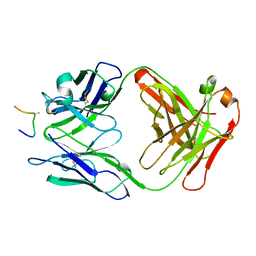 | | HIV-1 V3 LOOP MIMIC | | Descriptor: | AIB142, IGG1-KAPPA 59.1 FAB (HEAVY CHAIN), IGG1-KAPPA 59.1 FAB (LIGHT CHAIN) | | Authors: | Ghiara, J.B, Wilson, I.A. | | Deposit date: | 1996-11-06 | | Release date: | 1997-05-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of a constrained peptide mimic of the HIV-1 V3 loop neutralization site.
J.Mol.Biol., 266, 1997
|
|
1AI4
 
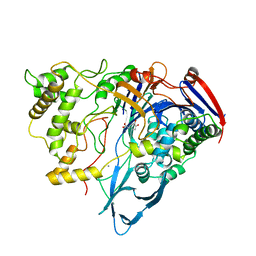 | |
1AI5
 
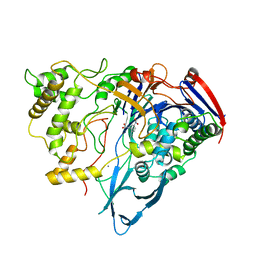 | | PENICILLIN ACYLASE COMPLEXED WITH M-NITROPHENYLACETIC ACID | | Descriptor: | 2-(3-NITROPHENYL)ACETIC ACID, CALCIUM ION, PENICILLIN AMIDOHYDROLASE | | Authors: | Done, S.H. | | Deposit date: | 1997-05-01 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Ligand-induced conformational change in penicillin acylase.
J.Mol.Biol., 284, 1998
|
|
1AI6
 
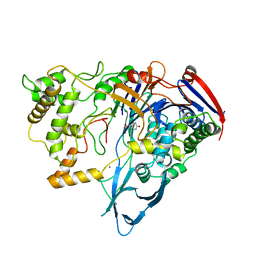 | | PENICILLIN ACYLASE WITH P-HYDROXYPHENYLACETIC ACID | | Descriptor: | 4-HYDROXYPHENYLACETATE, CALCIUM ION, PENICILLIN AMIDOHYDROLASE | | Authors: | Done, S.H. | | Deposit date: | 1997-05-01 | | Release date: | 1997-11-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Ligand-induced conformational change in penicillin acylase.
J.Mol.Biol., 284, 1998
|
|
1AI7
 
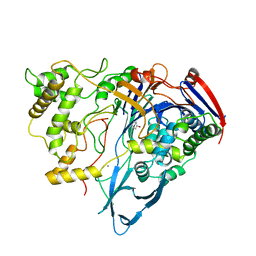 | | PENICILLIN ACYLASE COMPLEXED WITH PHENOL | | Descriptor: | CALCIUM ION, PENICILLIN AMIDOHYDROLASE, PHENOL | | Authors: | Done, S.H. | | Deposit date: | 1997-05-01 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand-induced conformational change in penicillin acylase.
J.Mol.Biol., 284, 1998
|
|
1AIE
 
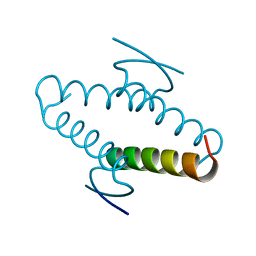 | |
1AII
 
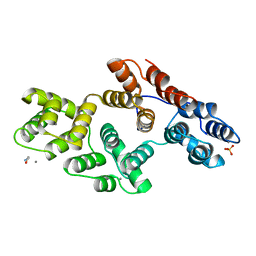 | | ANNEXIN III | | Descriptor: | ANNEXIN III, CALCIUM ION, ETHANOLAMINE, ... | | Authors: | Lewit-Bentley, A, Perron, B. | | Deposit date: | 1996-11-28 | | Release date: | 1997-03-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Can enzymatic activity, or otherwise, be inferred from structural studies of annexin III?
J.Biol.Chem., 272, 1997
|
|
1AIL
 
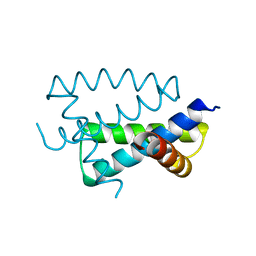 | | N-TERMINAL FRAGMENT OF NS1 PROTEIN FROM INFLUENZA A VIRUS | | Descriptor: | NONSTRUCTURAL PROTEIN NS1 | | Authors: | Liu, J, Lynch, P.A, Chien, C, Montelione, G.T, Krug, R.M, Berman, H.M. | | Deposit date: | 1997-04-21 | | Release date: | 1997-10-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the unique RNA-binding domain of the influenza virus NS1 protein.
Nat.Struct.Biol., 4, 1997
|
|
1AIP
 
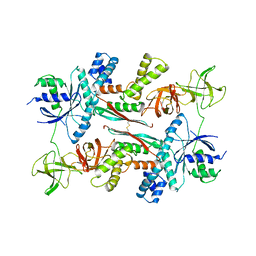 | | EF-TU EF-TS COMPLEX FROM THERMUS THERMOPHILUS | | Descriptor: | ELONGATION FACTOR TS, ELONGATION FACTOR TU | | Authors: | Wang, Y, Jiang, Y, Meyering-Voss, M, Sprinzl, M, Sigler, P.B. | | Deposit date: | 1997-04-22 | | Release date: | 1997-10-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the EF-Tu.EF-Ts complex from Thermus thermophilus.
Nat.Struct.Biol., 4, 1997
|
|
1AIS
 
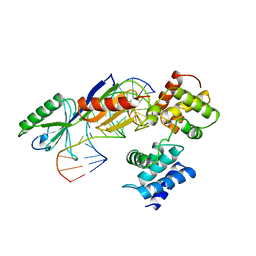 | | TATA-BINDING PROTEIN/TRANSCRIPTION FACTOR (II)B/TATA-BOX COMPLEX FROM PYROCOCCUS WOESEI | | Descriptor: | DNA (5'-D(*AP*AP*CP*TP*TP*AP*CP*TP*TP*TP*(5IU)P*(5IU)P*AP*AP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*TP*TP*AP*AP*AP*AP*AP*GP*TP*AP*AP*GP*TP*T )-3'), PROTEIN (TATA-BINDING PROTEIN), ... | | Authors: | Kosa, P.F, Ghosh, G, Dedecker, B.S, Sigler, P.B. | | Deposit date: | 1997-04-24 | | Release date: | 1997-07-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1-A crystal structure of an archaeal preinitiation complex: TATA-box-binding protein/transcription factor (II)B core/TATA-box.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1AIZ
 
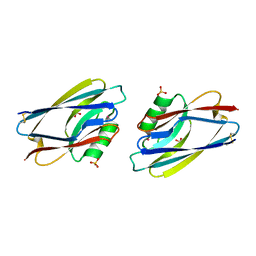 | | STRUCTURE OF APO-AZURIN FROM ALCALIGENES DENITRIFICANS AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | AZURIN, CADMIUM ION, SULFATE ION | | Authors: | Baker, E.N, Anderson, B.F, Blackwell, K.A. | | Deposit date: | 1993-11-11 | | Release date: | 1994-01-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of apo-azurin from Alcaligenes denitrificans at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1AJ5
 
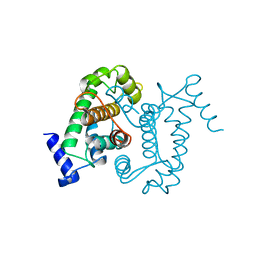 | | CALPAIN DOMAIN VI APO | | Descriptor: | CALPAIN | | Authors: | Cygler, M, Grochulski, P, Blanchard, H. | | Deposit date: | 1997-05-15 | | Release date: | 1998-05-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a calpain Ca(2+)-binding domain reveals a novel EF-hand and Ca(2+)-induced conformational changes.
Nat.Struct.Biol., 4, 1997
|
|
1AJ6
 
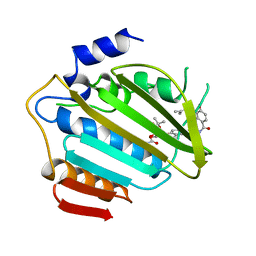 | |
1AJ7
 
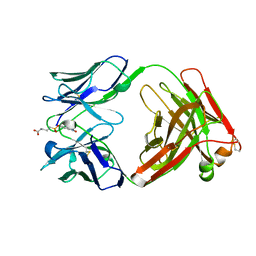 | | IMMUNOGLOBULIN 48G7 GERMLINE FAB ANTIBODY COMPLEXED WITH HAPTEN 5-(PARA-NITROPHENYL PHOSPHONATE)-PENTANOIC ACID. AFFINITY MATURATION OF AN ESTEROLYTIC ANTIBODY | | Descriptor: | 5-(PARA-NITROPHENYL PHOSPHONATE)-PENTANOIC ACID, IMMUNOGLOBULIN 48G7 FAB (HEAVY CHAIN), IMMUNOGLOBULIN 48G7 FAB (LIGHT CHAIN) | | Authors: | Wedemayer, G.J, Wang, L.H, Patten, P.A, Schultz, P.G, Stevens, R.C. | | Deposit date: | 1997-05-15 | | Release date: | 1997-11-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the evolution of an antibody combining site.
Science, 276, 1997
|
|
1AJ9
 
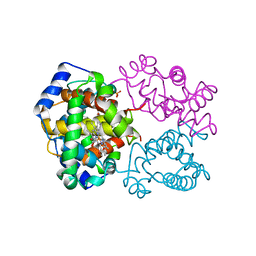 | | R-STATE HUMAN CARBONMONOXYHEMOGLOBIN ALPHA-A53S | | Descriptor: | CARBON MONOXIDE, HEMOGLOBIN (ALPHA CHAIN), HEMOGLOBIN (BETA CHAIN), ... | | Authors: | Vasquez, G.B, Ji, X, Fronticelli, C, Gilliland, G.L. | | Deposit date: | 1997-05-16 | | Release date: | 1998-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human carboxyhemoglobin at 2.2 A resolution: structure and solvent comparisons of R-state, R2-state and T-state hemoglobins.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1AJG
 
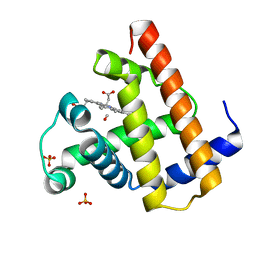 | | CARBONMONOXY MYOGLOBIN AT 40 K | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Teng, T.Y, Srajer, V, Moffat, K. | | Deposit date: | 1997-05-02 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Photolysis-induced structural changes in single crystals of carbonmonoxy myoglobin at 40 K.
Nat.Struct.Biol., 1, 1994
|
|
1AJH
 
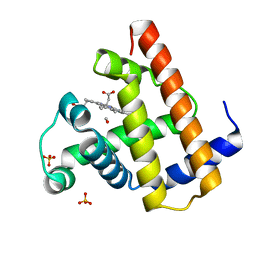 | | PHOTOPRODUCT OF CARBONMONOXY MYOGLOBIN AT 40 K | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Teng, T.Y, Srajer, V, Moffat, K. | | Deposit date: | 1997-05-02 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Photolysis-induced structural changes in single crystals of carbonmonoxy myoglobin at 40 K.
Nat.Struct.Biol., 1, 1994
|
|
1AJK
 
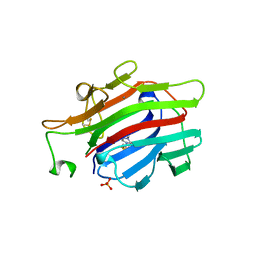 | | CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE CPA16M-84 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE, ... | | Authors: | Ay, J, Heinemann, U. | | Deposit date: | 1997-05-06 | | Release date: | 1998-05-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures and properties of de novo circularly permuted 1,3-1,4-beta-glucanases.
Proteins, 30, 1998
|
|
1AJN
 
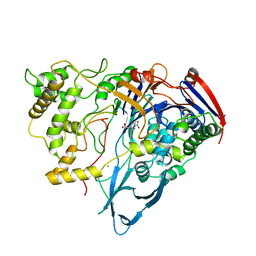 | | PENICILLIN ACYLASE COMPLEXED WITH P-NITROPHENYLACETIC ACID | | Descriptor: | 2-(4-NITROPHENYL)ACETIC ACID, CALCIUM ION, PENICILLIN AMIDOHYDROLASE | | Authors: | Done, S.H. | | Deposit date: | 1997-05-07 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Ligand-induced conformational change in penicillin acylase.
J.Mol.Biol., 284, 1998
|
|
1AJO
 
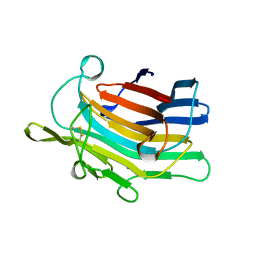 | | CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE CPA16M-127 | | Descriptor: | CALCIUM ION, CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE CPA16M-127 | | Authors: | Ay, J, Heinemann, U. | | Deposit date: | 1997-05-07 | | Release date: | 1998-05-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structures and properties of de novo circularly permuted 1,3-1,4-beta-glucanases.
Proteins, 30, 1998
|
|
1AJP
 
 | |
