1GKZ
 
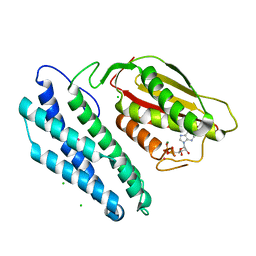 | | Branched-chain alpha-ketoacid dehydrogenase kinase (BCK) complxed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Machius, M, Chuang, J.L, Wynn, R.M, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2001-08-22 | | Release date: | 2001-10-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Rat Bckd Kinase: Nucleotide-Induced Domain Communication in a Mitochondrial Protein Kinase.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
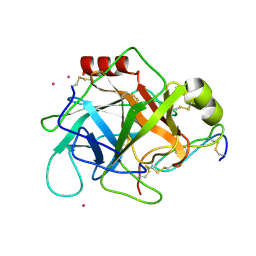
1GL0
 
 | |
1GL1
 
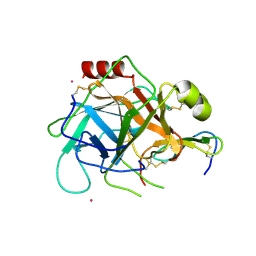 | |
1GL3
 
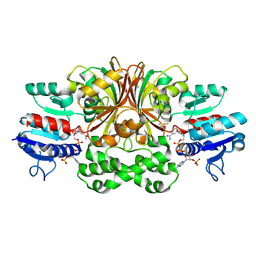 | | ASPARTATE BETA-SEMIALDEHYDE DEHYDROGENASE IN COMPLEX WITH NADP AND SUBSTRATE ANALOGUE S-METHYL CYSTEINE SULFOXIDE | | Descriptor: | ASPARTATE-SEMIALDEHYDE DEHYDROGENASE, CYSTEINE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hadfield, A.T, Kryger, G, Ouyang, J, Ringe, D, Petsko, G.A, Viola, R.E. | | Deposit date: | 2001-08-23 | | Release date: | 2001-11-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Active Site Analysis of the Potential Antimicrobial Target Aspartate Semialdehyde Dehydrogenase.
Biochemistry, 40, 2001
|
|
1GL4
 
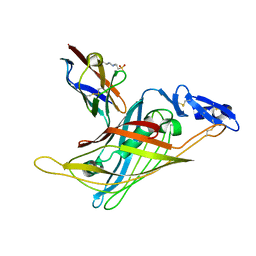 | | Nidogen-1 G2/Perlecan IG3 Complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BASEMENT MEMBRANE-SPECIFIC HEPARAN SULFATE PROTEOGLYCAN CORE PROTEIN, NIDOGEN-1, ... | | Authors: | Kvansakul, M, Hopf, M, Ries, A, Timpl, R, Hohenester, E. | | Deposit date: | 2001-08-23 | | Release date: | 2001-11-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the High-Affinity Interaction of Nidogen-1 with Immunoglobulin-Like Domain 3 of Perlecan
Embo J., 20, 2001
|
|
1GL9
 
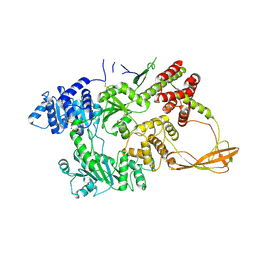 | |
1GLF
 
 | | CRYSTAL STRUCTURES OF ESCHERICHIA COLI GLYCEROL KINASE AND THE MUTANT A65T IN AN INACTIVE TETRAMER: CONFORMATIONAL CHANGES AND IMPLICATIONS FOR ALLOSTERIC REGULATION | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Feese, M.D, Faber, H.R, Bystrom, C.E, Pettigrew, D.W, Remington, S.J. | | Deposit date: | 1998-08-30 | | Release date: | 1998-10-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Glycerol kinase from Escherichia coli and an Ala65-->Thr mutant: the crystal structures reveal conformational changes with implications for allosteric regulation.
Structure, 6, 1998
|
|
1GLI
 
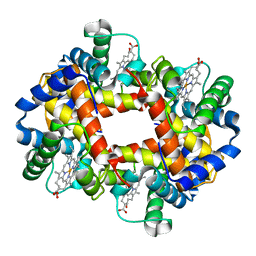 | |
1GLJ
 
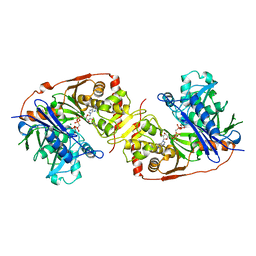 | | ESCHERICHIA COLI GLYCEROL KINASE MUTANT WITH BOUND ATP ANALOG SHOWING SUBSTANTIAL DOMAIN MOTION | | Descriptor: | GAMMA-ARSONO-BETA, GAMMA-METHYLENEADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Bystrom, C.E, Pettigrew, D.W, Branchaud, B.P, Remington, S.J. | | Deposit date: | 1998-09-03 | | Release date: | 1999-05-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of Escherichia coli glycerol kinase variant S58-->W in complex with nonhydrolyzable ATP analogues reveal a putative active conformation of the enzyme as a result of domain motion.
Biochemistry, 38, 1999
|
|
1GLL
 
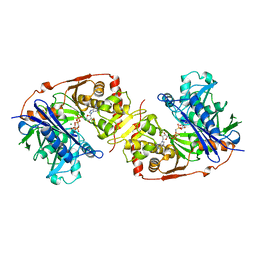 | | ESCHERICHIA COLI GLYCEROL KINASE MUTANT WITH BOUND ATP ANALOG SHOWING SUBSTANTIAL DOMAIN MOTION | | Descriptor: | GLYCEROL, GLYCEROL KINASE, MAGNESIUM ION, ... | | Authors: | Bystrom, C.E, Pettigrew, D.W, Branchaud, B.P, Remington, S.J. | | Deposit date: | 1998-09-24 | | Release date: | 1999-05-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of Escherichia coli glycerol kinase variant S58-->W in complex with nonhydrolyzable ATP analogues reveal a putative active conformation of the enzyme as a result of domain motion.
Biochemistry, 38, 1999
|
|
1GM4
 
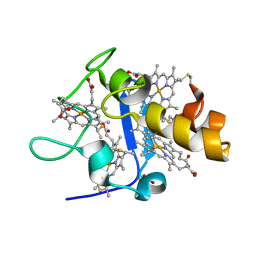 | | OXIDISED STRUCTURE OF CYTOCHROME C3 FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 at pH 7.6 | | Descriptor: | CYTOCHROME C3, HEME C, SULFATE ION | | Authors: | Bento, I, Louro, R, Matias, P.M, Catarino, T, Baptista, A.M, Soares, C.M, Carrondo, M.A, Turner, D.L, Xavier, A.V. | | Deposit date: | 2001-09-10 | | Release date: | 2002-09-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformational Component in the Coupled Transfer of Multiple Electrons and Protons in a Monomeric Tetraheme Cytochrome.
J.Biol.Chem., 276, 2001
|
|
1GM5
 
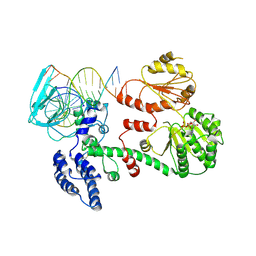 | | Structure of RecG bound to three-way DNA junction | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-(*CP*AP*GP*CP*TP*CP*CP*AP*TP*GP*AP*TP* CP*AP*TP*TP*GP*GP*CP*A)-3'), DNA (5'-(*GP*AP*GP*CP*AP*CP*TP*GP*C)-3'), ... | | Authors: | Singleton, M.R, Scaife, S, Wigley, D.B. | | Deposit date: | 2001-09-11 | | Release date: | 2001-10-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Structural Analysis of DNA Replication Fork Reversal by Recg
Cell(Cambridge,Mass.), 107, 2001
|
|
1GM6
 
 | | 3-D STRUCTURE OF A SALIVARY LIPOCALIN FROM BOAR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, GLYCEROL, ... | | Authors: | Spinelli, S, Vincent, F, Pelosi, P, Tegoni, M, Cambillau, C. | | Deposit date: | 2001-09-11 | | Release date: | 2002-05-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Boar Salivary Lipocalin. Three-Dimensional X-Ray Structure and Androsterol/Androstenone Docking Simulations.
Eur.J.Biochem., 269, 2002
|
|
1GM7
 
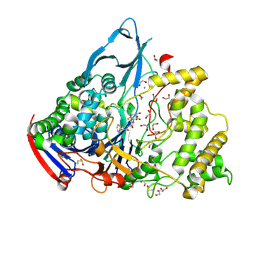 | | Crystal structures of penicillin acylase enzyme-substrate complexes: Structural insights into the catalytic mechanism | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PENICILLIN G, ... | | Authors: | McVey, C.E, Walsh, M.A, Dodson, G.G, Wilson, K.S, Brannigan, J.A. | | Deposit date: | 2001-09-11 | | Release date: | 2001-11-28 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structures of Penicillin Acylase Enzyme- Substrate Complexes: Structural Insights Into the Catalytic Mechanism
J.Mol.Biol., 313, 2001
|
|
1GM8
 
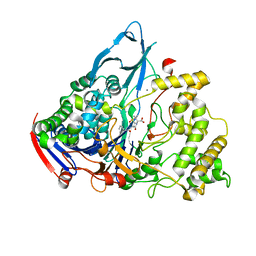 | | Crystal structures of penicillin acylase enzyme-substrate complexes: Structural insights into the catalytic mechanism | | Descriptor: | CALCIUM ION, N-[(2S,4S,6R)-2-(DIHYDROXYMETHYL)-4-HYDROXY-3,3-DIMETHYL-7-OXO-4LAMBDA~4~-THIA-1-AZABICYCLO[3.2.0]HEPT-6-YL]-2-PHENYLAC ETAMIDE, PENICILLIN G ACYLASE ALPHA SUBUNIT, ... | | Authors: | McVey, C.E, Walsh, M.A, Dodson, G.G, Wilson, K.S, Brannigan, J.A. | | Deposit date: | 2001-09-11 | | Release date: | 2001-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Penicillin Acylase Enzyme- Substrate Complexes: Structural Insights Into the Catalytic Mechanism
J.Mol.Biol., 313, 2001
|
|
1GM9
 
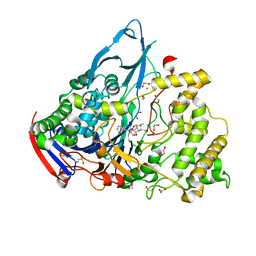 | | Crystal structures of penicillin acylase enzyme-substrate complexes: Structural insights into the catalytic mechanism | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, N-[(2S,4S,6R)-2-(DIHYDROXYMETHYL)-4-HYDROXY-3,3-DIMETHYL-7-OXO-4LAMBDA~4~-THIA-1-AZABICYCLO[3.2.0]HEPT-6-YL]-2-PHENYLAC ETAMIDE, ... | | Authors: | McVey, C.E, Walsh, M.A, Dodson, G.G, Wilson, K.S, Brannigan, J.A. | | Deposit date: | 2001-09-12 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Penicillin Acylase Enzyme- Substrate Complexes: Structural Insights Into the Catalytic Mechanism
J.Mol.Biol., 313, 2001
|
|
1GMB
 
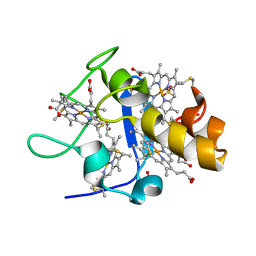 | | Reduced structure of CYTOCHROME C3 FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 at pH 7.6 | | Descriptor: | CYTOCHROME C3, HEME C, SULFATE ION | | Authors: | Bento, I, Louro, R, Matias, P.M, Catarino, T, Baptista, A.M, Soares, C.M, Carrondo, M.A, Turner, D.L, Xavier, A.V. | | Deposit date: | 2001-09-12 | | Release date: | 2002-09-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Component in the Coupled Transfer of Multiple Electrons and Protons in a Monomeric Tetraheme Cytochrome
J.Biol.Chem., 276, 2001
|
|
1GME
 
 | | Crystal structure and assembly of an eukaryotic small heat shock protein | | Descriptor: | HEAT SHOCK PROTEIN 16.9B | | Authors: | Van Montfort, R.L.M, Basha, E, Friedrich, K.L, Slingsby, C, Vierling, E. | | Deposit date: | 2001-09-13 | | Release date: | 2001-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure and Assembly of an Eukaryotic Small Heat Shock Protein
Nat.Struct.Biol., 8, 2001
|
|
1GMG
 
 | |
1GMI
 
 | | Structure of the c2 domain from novel protein kinase C epsilon | | Descriptor: | MAGNESIUM ION, PROTEIN KINASE C, EPSILON TYPE | | Authors: | Ochoa, W.F, Garcia-Garcia, J, Fita, I, Corbalan-Garcia, S, Verdaguer, N, Gomez-Fernandez, J.C. | | Deposit date: | 2001-09-14 | | Release date: | 2001-10-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the C2 Domain from Novel Protein Kinase Cepsilon. A Membrane Binding Model for Ca(2+ )-Independent C2 Domains
J.Mol.Biol., 311, 2001
|
|
1GMJ
 
 | | The structure of bovine IF1, the regulatory subunit of mitochondrial F-ATPase | | Descriptor: | ATPASE INHIBITOR | | Authors: | Cabezon, E, Runswick, M.J, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2001-09-14 | | Release date: | 2002-01-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of Bovine If(1), the Regulatory Subunit of Mitochondrial F-ATPase.
Embo J., 20, 2001
|
|
1GML
 
 | | crystal structure of the mouse CCT gamma apical domain (triclinic) | | Descriptor: | GLYCEROL, T-COMPLEX PROTEIN 1 SUBUNIT GAMMA | | Authors: | Pappenberger, G, Wilsher, J.A, Roe, S.M, Willison, K.R, Pearl, L.H. | | Deposit date: | 2001-09-17 | | Release date: | 2002-06-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Cct Gamma Apical Domain:: Implications for Substrate Binding to the Eukaryotic Cytosolic Chaperonin
J.Mol.Biol., 318, 2002
|
|
1GMM
 
 | | Carbohydrate binding module CBM6 from xylanase U Clostridium thermocellum | | Descriptor: | CALCIUM ION, CBM6, SODIUM ION, ... | | Authors: | Czjzek, M, Mosbah, A, Bolam, D, Allouch, J, Zamboni, V, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2001-09-19 | | Release date: | 2001-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Location of the Ligand-Binding Site of Carbohydrate-Binding Modules that Have Evolved from a Common Sequence is not Conserved.
J.Biol.Chem., 276, 2001
|
|
1GMN
 
 | | CRYSTAL STRUCTURES OF NK1-HEPARIN COMPLEXES REVEAL THE BASIS FOR NK1 ACTIVITY AND ENABLE ENGINEERING OF POTENT AGONISTS OF THE MET RECEPTOR | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEPATOCYTE GROWTH FACTOR | | Authors: | Lietha, D, Chirgadze, D.Y, Mulloy, B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2001-09-19 | | Release date: | 2001-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Nk1-Heparin Complexes Reveal the Basis for Nk1 Activity and Enable Engineering of Potent Agonists of the met Receptor
Embo J., 20, 2001
|
|
1GMO
 
 | | CRYSTAL STRUCTURES OF NK1-HEPARIN COMPLEXES REVEAL THE BASIS FOR NK1 ACTIVITY AND ENABLE ENGINEERING OF POTENT AGONISTS OF THE MET RECEPTOR | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, ... | | Authors: | Lietha, D, Chirgadze, D.Y, Mulloy, B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2001-09-20 | | Release date: | 2001-10-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of Nk1-Heparin Complexes Reveal the Basis for Nk1 Activity and Enable Engineering of Potent Agonists of the met Receptor
Embo J., 20, 2001
|
|
