1O8H
 
 | |
1O8I
 
 | |
1O8J
 
 | |
1O8K
 
 | |
1O8L
 
 | |
1O8M
 
 | |
1O8N
 
 | | The active site of the molybdenum cofactor biosynthetic protein domain Cnx1G | | Descriptor: | MOLYBDOPTERIN BIOSYNTHESIS CNX1 PROTEIN | | Authors: | Kuper, J, Winking, J, Hecht, H.J, Schwarz, G, Mendel, R.R. | | Deposit date: | 2002-11-28 | | Release date: | 2003-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Active Site of the Molybdenum Cofactor Biosynthetic Protein Domain Cnx1G
Arch.Biochem.Biophys., 411, 2003
|
|
1O8O
 
 | | The active site of the molybdenum cofactor biosynthetic protein domain Cnx1G | | Descriptor: | MOLYBDOPTERIN BIOSYNTHESIS CNX1 PROTEIN | | Authors: | Kuper, J, Winking, J, Hecht, H.J, Schwarz, G, Mendel, R.R. | | Deposit date: | 2002-11-28 | | Release date: | 2003-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Active Site of the Molybdenum Cofactor Biosynthetic Protein Domain Cnx1G
Arch.Biochem.Biophys., 411, 2003
|
|
1O8P
 
 | | Unbound structure of CsCBM6-3 from Clostridium stercorarium | | Descriptor: | CALCIUM ION, PUTATUVE ENDO-XYLANASE | | Authors: | Boraston, A.B, Notenboom, V, Warren, R.A.J, Kilbrun, D.G, Rose, D.R, Davies, G.J. | | Deposit date: | 2002-11-28 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and ligand binding of carbohydrate-binding module CsCBM6-3 reveals similarities with fucose-specific lectins and "galactose-binding" domains.
J. Mol. Biol., 327, 2003
|
|
1O8Q
 
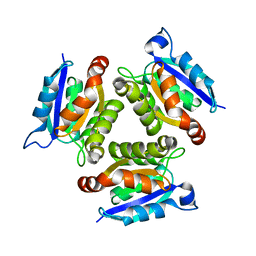 | | The active site of the molybdenum cofactor biosenthetic protein domain Cnx1G | | Descriptor: | MOLYBDOPTERIN BIOSYNTHESIS CNX1 PROTEIN | | Authors: | Kuper, J, Winking, J, Hecht, H.J, Schwarz, G, Mendel, R.R. | | Deposit date: | 2002-11-28 | | Release date: | 2003-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Active Site of the Molybdenum Cofactor Biosynthetic Protein Domain Cnx1G
Arch.Biochem.Biophys., 411, 2003
|
|
1O8S
 
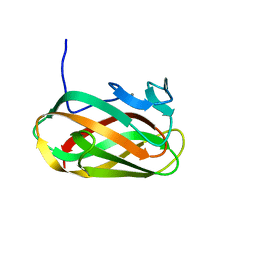 | | Structure of CsCBM6-3 from Clostridium stercorarium in complex with cellobiose | | Descriptor: | CALCIUM ION, PUTATIVE ENDO-XYLANASE, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Boraston, A.B, Notenboom, V, Warren, R.A.J, Kilbrun, D.G, Rose, D.R, Davies, G.J. | | Deposit date: | 2002-11-28 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure and Ligand Binding of Carbohydrate-Binding Module Cscbm6-3 Reveals Similarities with Fucose-Specific Lectins and Galactose-Binding Domains
J.Mol.Biol., 327, 2003
|
|
1O8V
 
 | | The crystal structure of Echinococcus granulosus fatty-acid-binding protein 1 | | Descriptor: | FATTY ACID BINDING PROTEIN HOMOLOG, PALMITIC ACID | | Authors: | Jakobsson, E, Alvite, G, Bergfors, T, Esteves, A, Kleywegt, G.J. | | Deposit date: | 2002-12-04 | | Release date: | 2003-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of Echinococcus Granulosus Fatty-Acid-Binding Protein 1
Biochim.Biophys.Acta, 1649, 2003
|
|
1O8X
 
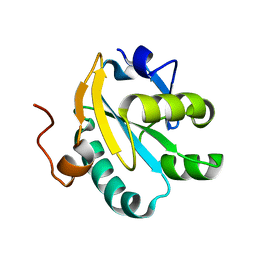 | | Mutant tryparedoxin-I Cys43Ala | | Descriptor: | TRYPAREDOXIN | | Authors: | Alphey, M.S, Bond, C.S, Hunter, W.N. | | Deposit date: | 2002-12-09 | | Release date: | 2003-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Tryparedoxins from Crithidia Fasciculata and Trypanosoma Brucei: Photoreduction of the Redox Disulfide Using Synchrotron Radiation and Evidence for a Conformational Switch Implicated in Function
J.Biol.Chem., 278, 2003
|
|
1O91
 
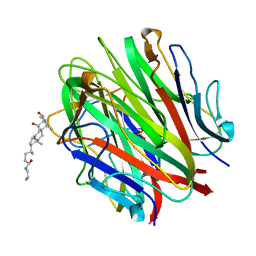 | | Crystal Structure of a Collagen VIII NC1 Domain Trimer | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, COLLAGEN ALPHA 1(VIII) CHAIN, SULFATE ION | | Authors: | Kvansakul, M, Bogin, O, Hohenester, E, Yayon, A. | | Deposit date: | 2002-12-10 | | Release date: | 2003-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Collagen Alpha1(Viii) Nc1 Trimer.
Matrix Biol., 22, 2003
|
|
1O94
 
 | | Ternary complex between trimethylamine dehydrogenase and electron transferring flavoprotein | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ELECTRON TRANSFER FLAVOPROTEIN ALPHA-SUBUNIT, ... | | Authors: | Leys, D, Basran, J, Talfournier, F, Sutcliffe, M.J, Scrutton, N.S. | | Deposit date: | 2002-12-11 | | Release date: | 2003-02-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Extensive Conformational Sampling in a Ternary Electron Transfer Complex.
Nat.Struct.Biol., 10, 2003
|
|
1O95
 
 | | Ternary complex between trimethylamine dehydrogenase and electron transferring flavoprotein | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ELECTRON TRANSFER FLAVOPROTEIN ALPHA-SUBUNIT, ... | | Authors: | Leys, D, Basran, J, Talfournier, F, Sutcliffe, M.J, Scrutton, N.S. | | Deposit date: | 2002-12-11 | | Release date: | 2003-02-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Extensive Conformational Sampling in a Ternary Electron Transfer Complex.
Nat.Struct.Biol., 10, 2003
|
|
1O96
 
 | | Structure of electron transferring flavoprotein for Methylophilus methylotrophus. | | Descriptor: | ADENOSINE MONOPHOSPHATE, ELECTRON TRANSFERRING FLAVOPROTEIN ALPHA-SUBUNIT, ELECTRON TRANSFERRING FLAVOPROTEIN BETA-SUBUNIT, ... | | Authors: | Leys, D, Basran, J, Talfournier, F, Sutcliffe, M.J, Scrutton, N.S. | | Deposit date: | 2002-12-11 | | Release date: | 2003-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Extensive Conformational Sampling in a Ternary Electron Transfer Complex.
Nat.Struct.Biol., 10, 2003
|
|
1O97
 
 | | Structure of electron transferring flavoprotein from Methylophilus methylotrophus, recognition loop removed by limited proteolysis | | Descriptor: | ADENOSINE MONOPHOSPHATE, ELECTRON TRANSFERRING FLAVOPROTEIN ALPHA-SUBUNIT, ELECTRON TRANSFERRING FLAVOPROTEIN BETA-SUBUNIT, ... | | Authors: | Leys, D, Basran, J, Talfournier, F, Sutcliffe, M.J, Scrutton, N.S. | | Deposit date: | 2002-12-11 | | Release date: | 2003-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Extensive Conformational Sampling in a Ternary Electron Transfer Complex.
Nat.Struct.Biol., 10, 2003
|
|
1O98
 
 | | 1.4A CRYSTAL STRUCTURE OF PHOSPHOGLYCERATE MUTASE FROM BACILLUS STEAROTHERMOPHILUS COMPLEXED WITH 2-PHOSPHOGLYCERATE | | Descriptor: | 2,3-BISPHOSPHOGLYCERATE-INDEPENDENT PHOSPHOGLYCERATE MUTASE, 2-PHOSPHOGLYCERIC ACID, MANGANESE (II) ION, ... | | Authors: | Rigden, D.J, Lamani, E, Littlejohn, J.E, Jedrzejas, M.J. | | Deposit date: | 2002-12-11 | | Release date: | 2003-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Insights Into the Catalytic Mechanism of Cofactor-Independent Phosphoglycerate Mutase from X-Ray Crystallography, Simulated Dynamics and Molecular Modeling
J.Mol.Biol., 328, 2003
|
|
1O99
 
 | | CRYSTAL STRUCTURE OF THE S62A MUTANT OF PHOSPHOGLYCERATE MUTASE FROM BACILLUS STEAROTHERMOPHILUS COMPLEXED WITH 2-PHOSPHOGLYCERATE | | Descriptor: | 2,3-BISPHOSPHOGLYCERATE-INDEPENDENT PHOSPHOGLYCERATE MUTASE, 2-PHOSPHOGLYCERIC ACID, MANGANESE (II) ION, ... | | Authors: | Rigden, D.J, Lamani, E, Littlejohn, J.E, Jedrzejas, M.J. | | Deposit date: | 2002-12-11 | | Release date: | 2002-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Insights Into the Catalytic Mechanism of Cofactor-Independent Phosphoglycerate Mutase from X-Ray Crystallography, Simulated Dynamics and Molecular Modeling
J.Mol.Biol., 328, 2003
|
|
1O9B
 
 | | QUINATE/SHIKIMATE DEHYDROGENASE YDIB COMPLEXED WITH NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, HYPOTHETICAL SHIKIMATE 5-DEHYDROGENASE-LIKE PROTEIN YDIB, PHOSPHATE ION | | Authors: | Michel, G, Cygler, M. | | Deposit date: | 2002-12-12 | | Release date: | 2003-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Shikimate Dehydrogenase Aroe and its Paralog Ydib: A Common Structural Framework for Different Activities
J.Biol.Chem., 278, 2003
|
|
1O9C
 
 | | Structural view of a fungal toxin acting on a 14-3-3 regulatory complex | | Descriptor: | 14-3-3-LIKE PROTEIN C, CHLORIDE ION, CITRATE ANION | | Authors: | Wurtele, M, Jelich-Ottmann, C, Wittinghofer, A, Oecking, C. | | Deposit date: | 2002-12-12 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural View of a Fungal Toxin Acting on a 14-3-3 Regulatory Complex
Embo J., 22, 2003
|
|
1O9D
 
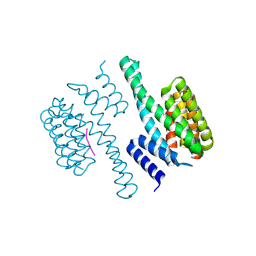 | | Structural view of a fungal toxin acting on a 14-3-3 regulatory complex | | Descriptor: | 14-3-3-LIKE PROTEIN C, PLASMA MEMBRANE H+ ATPASE | | Authors: | Wurtele, M, Jelich-Ottmann, C, Wittinghofer, A, Oecking, C. | | Deposit date: | 2002-12-12 | | Release date: | 2003-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural View of a Fungal Toxin Acting on a 14-3-3 Regulatory Complex
Embo J., 22, 2003
|
|
1O9E
 
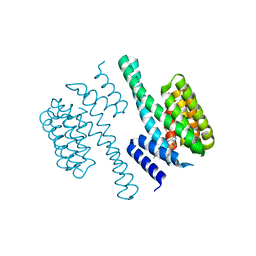 | | Structural view of a fungal toxin acting on a 14-3-3 regulatory complex | | Descriptor: | 14-3-3-LIKE PROTEIN C, CITRATE ANION, FUSICOCCIN | | Authors: | Wurtele, M, Jelich-Ottmann, C, Wittinghofer, A, Oecking, C. | | Deposit date: | 2002-12-12 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural View of a Fungal Toxin Acting on a 14-3-3 Regulatory Complex
Embo J., 22, 2003
|
|
1O9F
 
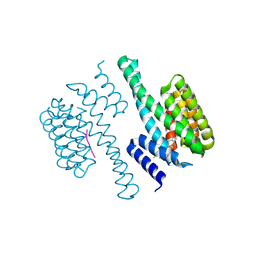 | | Structural view of a fungal toxin acting on a 14-3-3 regulatory complex | | Descriptor: | 14-3-3-LIKE PROTEIN C, FUSICOCCIN, PLASMA MEMBRANE H+ ATPASE | | Authors: | Wurtele, M, Jelich-Ottmann, C, Wittinghofer, A, Oecking, C. | | Deposit date: | 2002-12-12 | | Release date: | 2003-03-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural View of a Fungal Toxin Acting on a 14-3-3 Regulatory Complex
Embo J., 22, 2003
|
|
