1JV8
 
 | | NMR Structure of BPTI Mutant G37A | | Descriptor: | TRYPSIN INHIBITOR | | Authors: | Battiste, J.L, Li, R, Woodward, C. | | Deposit date: | 2001-08-28 | | Release date: | 2001-09-12 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | A highly destabilizing mutation, G37A, of the bovine pancreatic trypsin inhibitor retains the average native conformation but greatly increases local flexibility
Biochemistry, 41, 2002
|
|
1JV9
 
 | | NMR Structure of BPTI Mutant G37A | | Descriptor: | TRYPSIN INHIBITOR | | Authors: | Battiste, J.L, Li, R, Woodward, C. | | Deposit date: | 2001-08-28 | | Release date: | 2001-09-12 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | A highly destabilizing mutation, G37A, of the bovine pancreatic trypsin inhibitor retains the average native conformation but greatly increases local flexibility
Biochemistry, 41, 2002
|
|
1JVE
 
 | |
1JVR
 
 | | STRUCTURE OF THE HTLV-II MATRIX PROTEIN, NMR, 20 STRUCTURES | | Descriptor: | HUMAN T-CELL LEUKEMIA VIRUS TYPE II MATRIX PROTEIN | | Authors: | Christensen, A.M, Massiah, M.A, Turner, B.G, Sundquist, W.I, Summers, M.F. | | Deposit date: | 1996-10-03 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the HTLV-II matrix protein and comparative analysis of matrix proteins from the different classes of pathogenic human retroviruses.
J.Mol.Biol., 264, 1996
|
|
1JW2
 
 | | SOLUTION STRUCTURE OF HEMOLYSIN EXPRESSION MODULATING PROTEIN Hha FROM ESCHERICHIA COLI. Ontario Centre for Structural Proteomics target EC0308_1_72; Northeast Structural Genomics Target ET88 | | Descriptor: | HEMOLYSIN EXPRESSION MODULATING PROTEIN Hha | | Authors: | Chang, X, Yee, A, Savchenko, A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-09-02 | | Release date: | 2002-02-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JW3
 
 | | Solution Structure of Methanobacterium Thermoautotrophicum Protein 1598. Ontario Centre for Structural Proteomics target MTH1598_1_140; Northeast Structural Genomics Target TT6 | | Descriptor: | Conserved Hypothetical Protein MTH1598 | | Authors: | Chang, X, Connelly, G, Yee, A, Kennedy, M.A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-09-02 | | Release date: | 2002-02-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JWC
 
 | |
1JWD
 
 | |
1JWE
 
 | | NMR Structure of the N-Terminal Domain of E. Coli Dnab Helicase | | Descriptor: | PROTEIN (DNAB HELICASE) | | Authors: | Weigelt, J, Brown, S.E, Miles, C.S, Dixon, N.E, Otting, G. | | Deposit date: | 1999-01-22 | | Release date: | 1999-01-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the N-terminal domain of E. coli DnaB helicase: implications for structure rearrangements in the helicase hexamer.
Structure Fold.Des., 7, 1999
|
|
1JXC
 
 | | Minimized NMR structure of ATT, an Arabidopsis trypsin/chymotrypsin inhibitor | | Descriptor: | Putative trypsin inhibitor ATTI-2 | | Authors: | Zhao, Q, Chae, Y.K, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2001-09-06 | | Release date: | 2003-01-07 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of ATTp, an Arabidopsis thaliana Trypsin Inhibitor
Biochemistry, 41, 2002
|
|
1JY4
 
 | |
1JY6
 
 | |
1JY9
 
 | | MINIMIZED AVERAGE STRUCTURE OF DP-TT2 | | Descriptor: | DP-TT2 | | Authors: | Stanger, H.E, Syud, F.A, Espinosa, J.F, Giriat, I, Muir, T, Gellman, S.H. | | Deposit date: | 2001-09-11 | | Release date: | 2001-09-19 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Length-dependent stability and strand length limits in antiparallel beta -sheet secondary structure.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1JYT
 
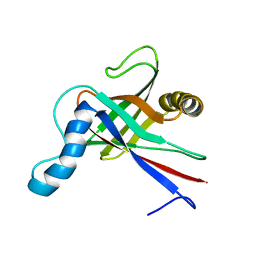 | | Solution structure of olfactory marker protein from rat | | Descriptor: | Olfactory Marker Protein | | Authors: | Baldisseri, D.M, Margolis, J.W, Weber, D.J, Koo, J.H, Margolis, F.M. | | Deposit date: | 2001-09-13 | | Release date: | 2001-10-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Olfactory marker protein (OMP) exhibits a beta-clam fold in solution: implications for target peptide interaction and olfactory signal transduction.
J.Mol.Biol., 319, 2002
|
|
1JZC
 
 | |
1JZP
 
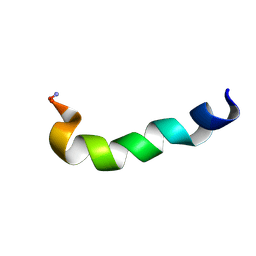 | | Modified Peptide A (D18-A1) of the Rabbit Skeletal Dihydropyridine Receptor | | Descriptor: | Skeletal Dihydropydrine Receptor | | Authors: | Green, D, Pace, S, Sakowska, M, Dulhunty, A.F, Casarotto, M.G. | | Deposit date: | 2001-09-17 | | Release date: | 2002-03-20 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structural surface of two beta-sheet scorpion toxins mimics that of an alpha-helical dihydropyridine receptor segment.
Biochem.J., 370, 2003
|
|
1JZU
 
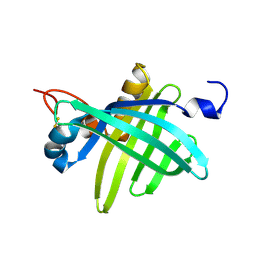 | | Cell transformation by the myc oncogene activates expression of a lipocalin: analysis of the gene (Q83) and solution structure of its protein product | | Descriptor: | lipocalin Q83 | | Authors: | Hartl, M, Matt, T, Schueler, W, Siemeister, G, Kontaxis, G, Kloiber, K, Konrat, R, Bister, K. | | Deposit date: | 2001-09-17 | | Release date: | 2003-07-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Cell Transformation by the v-myc Oncogene Abrogates c-Myc/Max-mediated Suppression of a
C/EBPbeta-dependent Lipocalin Gene.
J.Mol.Biol., 333, 2003
|
|
1K09
 
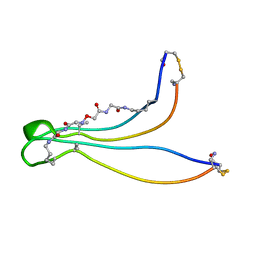 | | Solution structure of BetaCore, A Designed Water Soluble Four-Stranded Antiparallel b-sheet Protein | | Descriptor: | (7E)-4,9-dioxo-6-oxa-3,7,10-triazadodec-7-ene-1,12-dioic acid, Core Module I, Core Module II | | Authors: | Carulla, N, Woodward, C, Barany, G. | | Deposit date: | 2001-09-18 | | Release date: | 2002-07-10 | | Last modified: | 2024-02-21 | | Method: | SOLUTION NMR | | Cite: | BetaCore, a designed water soluble four-stranded antiparallel beta-sheet protein.
Protein Sci., 11, 2002
|
|
1K0P
 
 | |
1K0S
 
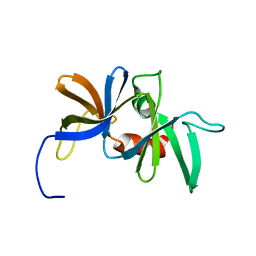 | | Solution structure of the chemotaxis protein CheW from the thermophilic organism Thermotoga maritima | | Descriptor: | CHEMOTAXIS PROTEIN CHEW | | Authors: | Griswold, I.J, Zhou, H, Swanson, R.V, Simon, M.I, Dahlquist, F.W. | | Deposit date: | 2001-09-20 | | Release date: | 2002-02-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure and interactions of CheW from Thermotoga maritima.
Nat.Struct.Biol., 9, 2002
|
|
1K0T
 
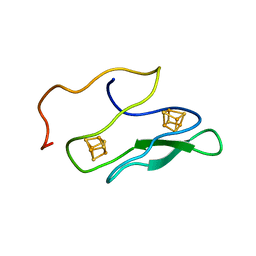 | | NMR SOLUTION STRUCTURE OF UNBOUND, OXIDIZED PHOTOSYSTEM I SUBUNIT PSAC, CONTAINING [4FE-4S] CLUSTERS FA AND FB | | Descriptor: | IRON/SULFUR CLUSTER, PSAC SUBUNIT OF PHOTOSYSTEM I | | Authors: | Antonkine, M.L, Liu, G, Bentrop, D, Bryant, D.A, Bertini, I, Luchinat, C, Golbeck, J.H, Stehlik, D. | | Deposit date: | 2001-09-20 | | Release date: | 2002-06-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the unbound, oxidized Photosystem I subunit PsaC, containing [4Fe-4S] clusters F(A) and F(B): a conformational change occurs upon binding to photosystem I.
J.Biol.Inorg.Chem., 7, 2002
|
|
1K0X
 
 | |
1K18
 
 | |
1K19
 
 | | NMR Solution Structure of the Chemosensory Protein CSP2 from Moth Mamestra brassicae | | Descriptor: | Chemosensory Protein CSP2 | | Authors: | Mosbah, A, Campanacci, V, Lartigue, A, Tegoni, M, Cambillau, C, Darbon, H. | | Deposit date: | 2001-09-24 | | Release date: | 2002-12-04 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a chemosensory protein from the moth Mamestra brassicae
BIOCHEM.J., 369, 2003
|
|
1K1C
 
 | | Solution Structure of Crh, the Bacillus subtilis Catabolite Repression HPr | | Descriptor: | catabolite repression HPr-like protein | | Authors: | Favier, A, Brutscher, B, Blackledge, M, Galinier, A, Deutscher, J, Penin, F, Marion, D. | | Deposit date: | 2001-09-25 | | Release date: | 2001-10-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of Crh, the Bacillus subtilis catabolite repression HPr.
J.Mol.Biol., 317, 2002
|
|
