2N5C
 
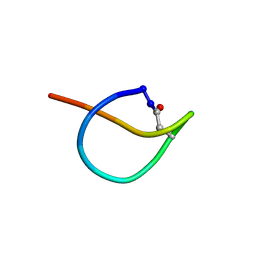 | | Solution NMR structure of the lasso peptide chaxapeptin | | Descriptor: | chaxapeptin | | Authors: | Elsayed, S.S, Trusch, F, Deng, H, Raab, A, Prokes, I, Busarakam, K, Asenjo, J.A, Andrews, B.A, van West, P, Bull, A.T, Goodfellow, M, Yi, Y, Ebel, R, Jaspars, M, Rateb, M.E. | | Deposit date: | 2015-07-14 | | Release date: | 2015-10-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Chaxapeptin, a Lasso Peptide from Extremotolerant Streptomyces leeuwenhoekii Strain C58 from the Hyperarid Atacama Desert.
J.Org.Chem., 80, 2015
|
|
2N5D
 
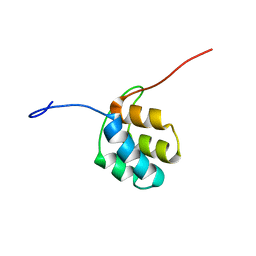 | | NMR structure of PKS domains | | Descriptor: | fusion protein of two PKS domains | | Authors: | Dorival, J, Annaval, T, Risser, F, Collin, S, Roblin, P, Jacob, C, Gruez, A, Chagot, B, Weissman, K.J. | | Deposit date: | 2015-07-14 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Characterization of Intersubunit Communication in the Virginiamycin trans-Acyl Transferase Polyketide Synthase.
J.Am.Chem.Soc., 138, 2016
|
|
2N5E
 
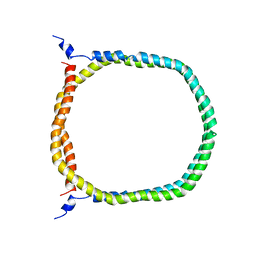 | | The 3D solution structure of discoidal high-density lipoprotein particles | | Descriptor: | Apolipoprotein A-I | | Authors: | Bibow, S, Polyhach, Y, Eichmann, C, Chi, C.N, Kowal, J, Stahlberg, H, Jeschke, G, Guentert, P, Riek, R. | | Deposit date: | 2015-07-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of discoidal high-density lipoprotein particles with a shortened apolipoprotein A-I.
Nat.Struct.Mol.Biol., 24, 2017
|
|
2N5F
 
 | |
2N5G
 
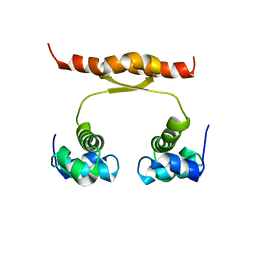 | | NMR structure of KorA, a plasmid-encoded, global transcription regulator KorA | | Descriptor: | TrfB transcriptional repressor protein | | Authors: | Rajasekar, K.V, Lovering, A.L, Dancea, F.V, Scott, D.J, Harris, S, Bingle, L.E, Roessle, M, Thomas, C.M, Hyde, E.I, White, S.A. | | Deposit date: | 2015-07-17 | | Release date: | 2016-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Flexibility of KorA, a plasmid-encoded, global transcription regulator, in the presence and the absence of its operator.
Nucleic Acids Res., 44, 2016
|
|
2N5H
 
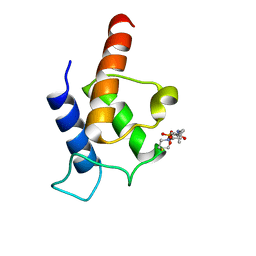 | | PltL-holo | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Peptidyl carrier protein PltL | | Authors: | Jaremko, M.J, Lee, D.J, Burkart, M.D. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and Substrate Sequestration in the Pyoluteorin Type II Peptidyl Carrier Protein PltL.
J.Am.Chem.Soc., 137, 2015
|
|
2N5I
 
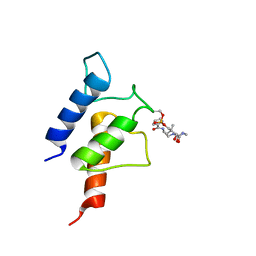 | | PltL-pyrrolyl | | Descriptor: | (3R)-3-hydroxy-2,2-dimethyl-4-oxo-4-{[3-oxo-3-({2-[(1H-pyrrol-2-ylcarbonyl)amino]ethyl}amino)propyl]amino}butyl dihydrogen phosphate, Peptidyl carrier protein PltL | | Authors: | Jaremko, M.J, Lee, D.J, Burkart, M.D. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and Substrate Sequestration in the Pyoluteorin Type II Peptidyl Carrier Protein PltL.
J.Am.Chem.Soc., 137, 2015
|
|
2N5J
 
 | | Regnase-1 N-terminal domain | | Descriptor: | Ribonuclease ZC3H12A | | Authors: | Yokogawa, M, Tsushima, T, Noda, N.N, Kumeta, H, Adachi, W, Enokizono, Y, Yamashita, K, Standley, D.M, Takeuchi, O, Akira, S, Inagaki, F. | | Deposit date: | 2015-07-18 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the regulation of enzymatic activity of Regnase-1 by domain-domain interactions
Sci Rep, 6, 2016
|
|
2N5K
 
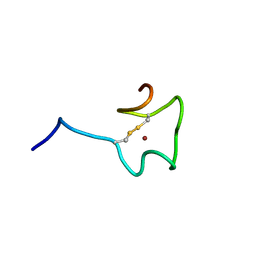 | | Regnase-1 Zinc finger domain | | Descriptor: | Ribonuclease ZC3H12A, ZINC ION | | Authors: | Yokogawa, M, Tsushima, T, Noda, N.N, Kumeta, H, Adachi, W, Enokizono, Y, Yamashita, K, Standley, D.M, Takeuchi, O, Akira, S, Inagaki, F. | | Deposit date: | 2015-07-18 | | Release date: | 2016-03-16 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the regulation of enzymatic activity of Regnase-1 by domain-domain interactions
Sci Rep, 6, 2016
|
|
2N5L
 
 | | Regnase-1 C-terminal domain | | Descriptor: | Ribonuclease ZC3H12A | | Authors: | Yokogawa, M, Tsushima, T, Noda, N.N, Kumeta, H, Adachi, W, Enokizono, Y, Yamashita, K, Standley, D.M, Takeuchi, O, Akira, S, Inagaki, F. | | Deposit date: | 2015-07-18 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the regulation of enzymatic activity of Regnase-1 by domain-domain interactions
Sci Rep, 6, 2016
|
|
2N5M
 
 | | Unveiling the structural determinants of KIAA0323 binding preference for NEDD8 | | Descriptor: | Protein KHNYN | | Authors: | Santonico, E, Nepravishta, R, Mattioni, A, Valentini, E, Mandaliti, W, Procopio, R, Iannuccelli, M, Castagnoli, L, Polo, S, Paci, M, Cesareni, G. | | Deposit date: | 2015-07-21 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Unveiling the structural determinants of KIAA0323 binding preference for NEDD8.
To be Published
|
|
2N5N
 
 | | Structure of an N-terminal domain of CHD4 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 4 | | Authors: | Silva, A.P.G, Mackay, J.P. | | Deposit date: | 2015-07-22 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The N-terminal Region of Chromodomain Helicase DNA-binding Protein 4 (CHD4) Is Essential for Activity and Contains a High Mobility Group (HMG) Box-like-domain That Can Bind Poly(ADP-ribose).
J.Biol.Chem., 291, 2016
|
|
2N5O
 
 | | Universal Base oligonucleotide structure | | Descriptor: | DNA_(5'-D(*AP*TP*GP*GP*(4EN)P*GP*CP*TP*C)-3'), DNA_(5'-D(*GP*AP*GP*CP*TP*CP*CP*AP*T)-3') | | Authors: | Spring-Connell, A.M, Evich, M.G, Seela, F, Germann, M.W. | | Deposit date: | 2015-07-23 | | Release date: | 2016-09-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Using NMR and molecular dynamics to link structure and dynamics effects of the universal base 8-aza, 7-deaza, N8 linked adenosine analog.
Nucleic Acids Res., 44, 2016
|
|
2N5P
 
 | | Universal base control oligonucleotide structure | | Descriptor: | DNA_(5'-D(*AP*TP*GP*GP*AP*GP*CP*TP*C)-3'), DNA_(5'-D(*GP*AP*GP*CP*TP*CP*CP*AP*T)-3') | | Authors: | Spring-Connell, A.M, Evich, M.G, Seela, F, Germann, M.W. | | Deposit date: | 2015-07-23 | | Release date: | 2016-09-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Using NMR and molecular dynamics to link structure and dynamics effects of the universal base 8-aza, 7-deaza, N8 linked adenosine analog.
Nucleic Acids Res., 44, 2016
|
|
2N5Q
 
 | | Solution structure of cystein-rich peptide jS1 from Jasminum sambac | | Descriptor: | cysteine-rich peptide jS1 | | Authors: | Shin, J, Kumari, G, Serra, A, Nguyen, P.Q.T, Yoon, H, Tam, J.P. | | Deposit date: | 2015-07-24 | | Release date: | 2016-07-27 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Identification of a cysteine-rich peptide family with unusual disulfide connectivity from Jasminum sambac
To be Published
|
|
2N5R
 
 | |
2N5S
 
 | |
2N5T
 
 | | Ensemble solution structure of the phosphoenolpyruvate-Enzyme I complex from the bacterial phosphotransferase system | | Descriptor: | Phosphoenolpyruvate-protein phosphotransferase | | Authors: | Venditti, V, Schwieters, C.D, Grishaev, A, Clore, G. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Dynamic equilibrium between closed and partially closed states of the bacterial Enzyme I unveiled by solution NMR and X-ray scattering.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2N5U
 
 | |
2N5W
 
 | | The NMR solution structure of octyl-tridecaptin A1 in DPC micelles | | Descriptor: | Octyl-tridecaptin A1 | | Authors: | Cochrane, S.A, Findlay, B, Bakhtiary, A, Acedo, J.Z, Rodriguez-Lopez, E.M, Vederas, J.C. | | Deposit date: | 2015-08-01 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Antimicrobial lipopeptide tridecaptin A1 selectively binds to Gram-negative lipid II.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2N5X
 
 | |
2N5Y
 
 | | Solution NMR structure of octyl-tridecaptin A1 in DPC micelles containing Gram-negative lipid II | | Descriptor: | Octyl-tridecaptin A1 | | Authors: | Cochrane, S.A, Findlay, B, Bakhtiary, A, Rodriguez-Lopez, E.M, Vederas, J.C. | | Deposit date: | 2015-08-03 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Antimicrobial lipopeptide tridecaptin A1 selectively binds to Gram-negative lipid II.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2N5Z
 
 | |
2N60
 
 | | G-quadruplexes with (4n-1) guanines in the G-tetrad core: formation of a G-triad water complex and implication for small-molecule binding | | Descriptor: | DNA (5'-D(*TP*TP*GP*TP*GP*TP*GP*GP*GP*TP*GP*GP*GP*TP*GP*GP*GP*T)-3') | | Authors: | Heddi, B, Martin-Pintado, N, Serimbetov, Z, Kari, T.M, Phan, A.T. | | Deposit date: | 2015-08-08 | | Release date: | 2016-06-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | G-quadruplexes with (4n - 1) guanines in the G-tetrad core: formation of a G-triadwater complex and implication for small-molecule binding
Nucleic Acids Res., 44, 2016
|
|
2N62
 
 | | ddFLN5+110 | | Descriptor: | gelation factor, secretion monitor chimera | | Authors: | Cabrita, L.D, Cassaignau, A.M.E, Launay, H.M.M, Waudby, C.A, Camilloni, C, Robertson, A.L, Wang, X, Wlodarski, T, Wentink, A.S, Vendruscolo, M, Dobson, C.M, Christodoulou, J. | | Deposit date: | 2015-08-10 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A structural ensemble of a ribosome-nascent chain complex during cotranslational protein folding.
Nat.Struct.Mol.Biol., 23, 2016
|
|
