1VKT
 
 | | HUMAN INSULIN TWO DISULFIDE MODEL, NMR, 10 STRUCTURES | | Descriptor: | INSULIN | | Authors: | Hua, Q.X, Hu, S.Q, Frank, B.H, Jia, W.H, Chu, Y.C, Wang, S.H, Burke, G.T, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 1996-10-14 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Mapping the functional surface of insulin by design: structure and function of a novel A-chain analogue.
J.Mol.Biol., 264, 1996
|
|
1VM2
 
 | |
1VM3
 
 | |
1VM4
 
 | |
1VM5
 
 | |
1VMC
 
 | | STROMA CELL-DERIVED FACTOR-1ALPHA (SDF-1ALPHA) | | Descriptor: | Stromal cell-derived factor 1 | | Authors: | Gozansky, E.K, Clore, G.M. | | Deposit date: | 2004-09-20 | | Release date: | 2005-03-01 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Mapping the binding of the N-terminal extracellular tail of the CXCR4 receptor to stromal cell-derived factor-1alpha.
J.Mol.Biol., 345, 2005
|
|
1VND
 
 | | VND/NK-2 PROTEIN (HOMEODOMAIN), NMR | | Descriptor: | VND/NK-2 PROTEIN | | Authors: | Tsao, D.H.H, Gruschus, J.M, Wang, L.-H, Nirenberg, M, Ferretti, J.A. | | Deposit date: | 1996-05-22 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional solution structure of the NK-2 homeodomain from Drosophila.
J.Mol.Biol., 251, 1995
|
|
1VPU
 
 | |
1VQX
 
 | | ARRESTIN-BOUND NMR STRUCTURES OF THE PHOSPHORYLATED CARBOXY-TERMINAL DOMAIN OF RHODOPSIN, REFINED | | Descriptor: | RHODOPSIN | | Authors: | Kisselev, O.G, Downs, M.A, Mcdowell, J.H, Hargrave, P.A. | | Deposit date: | 2005-01-07 | | Release date: | 2005-01-18 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Conformational Changes in the Phosphorylated C-Terminal Domain of Rhodopsin During Rhodopsin Arrestin Interactions
J.Biol.Chem., 279, 2004
|
|
1VRC
 
 | |
1VRE
 
 | | SOLUTION STRUCTURE OF COMPONENT IV GLYCERA DIBRANCHIATA MONOMERIC HEMOGLOBIN-CO | | Descriptor: | CARBON MONOXIDE, PROTEIN (GLOBIN, MONOMERIC COMPONENT M-IV), ... | | Authors: | Volkman, B.F, Alam, S.L, Satterlee, J.D, Markley, J.L. | | Deposit date: | 1999-03-25 | | Release date: | 1999-04-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of component IV Glycera dibranchiata monomeric hemoglobin-CO.
Biochemistry, 37, 1998
|
|
1VRV
 
 | |
1VSQ
 
 | |
1VTP
 
 | |
1VTX
 
 | |
1VYX
 
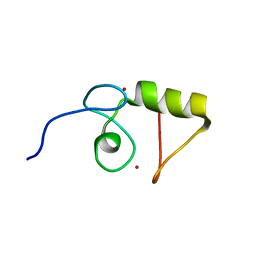 | | Solution structure of the KSHV K3 N-terminal domain | | Descriptor: | ORF K3, ZINC ION | | Authors: | Dodd, R.B, Allen, M.D, Brown, S.E, Sanderson, C.M, Duncan, L.M, lehner, P.J, Bycroft, M, Read, R.J. | | Deposit date: | 2004-05-07 | | Release date: | 2004-10-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Kaposi'S Sarcoma-Associated Herpesvirus K3 N-Terminal Domain Reveals a Novel E2-Binding C4Hc3-Type Ring Domain
J.Biol.Chem., 279, 2004
|
|
1VZS
 
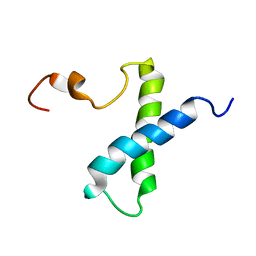 | | Solution structure of subunit F6 from the peripheral stalk region of ATP synthase from bovine heart mitochondria | | Descriptor: | ATP SYNTHASE COUPLING FACTOR 6, MITOCHONDRIAL PRECURSOR | | Authors: | Carbajo, R.J, Silvester, J.A, Runswick, M.J, Walker, J.E, Neuhaus, D. | | Deposit date: | 2004-05-25 | | Release date: | 2004-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Subunit F(6) from the Peripheral Stalk Region of ATP Synthase from Bovine Heart Mitochondria
J.Mol.Biol., 342, 2004
|
|
1W09
 
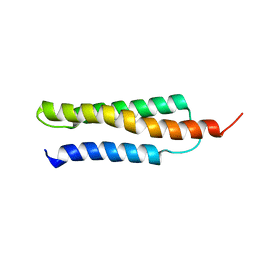 | | Solution structure of the cis form of the human alpha-hemoglobin stabilizing protein (AHSP) | | Descriptor: | ALPHA-HEMOGLOBIN STABILIZING PROTEIN | | Authors: | Santiveri, C.M, Perez-Canadillas, J.M, Vadivelu, M.K, Allen, M.D, Rutherford, T.J, Watkins, N.A, Bycroft, M. | | Deposit date: | 2004-05-25 | | Release date: | 2004-06-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the alpha-hemoglobin stabilizing protein: insights into conformational heterogeneity and binding.
J. Biol. Chem., 279, 2004
|
|
1W0A
 
 | | Solution structure of the trans form of the human alpha-hemoglobin stabilizing protein (AHSP) | | Descriptor: | ALPHA-HEMOGLOBIN STABILIZING PROTEIN | | Authors: | Santiveri, C.M, Perez-Canadillas, J.M, Vadivelu, M.K, Allen, M.D, Rutherford, T.J, Watkins, N.A, Bycroft, M. | | Deposit date: | 2004-06-01 | | Release date: | 2004-06-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the alpha-hemoglobin stabilizing protein: insights into conformational heterogeneity and binding.
J. Biol. Chem., 279, 2004
|
|
1W0B
 
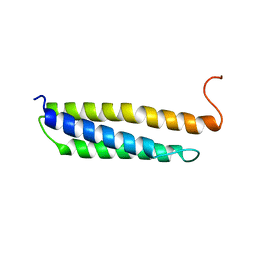 | | Solution structure of the human alpha-hemoglobin stabilizing protein (AHSP) P30A mutant | | Descriptor: | ALPHA-HEMOGLOBIN STABILIZING PROTEIN | | Authors: | Santiveri, C.M, Perez-Canadillas, J.M, Vadivelu, M.K, Allen, M.D, Rutherford, T.J, Watkins, N.A, Bycroft, M. | | Deposit date: | 2004-06-01 | | Release date: | 2004-06-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the alpha-hemoglobin stabilizing protein: insights into conformational heterogeneity and binding.
J. Biol. Chem., 279, 2004
|
|
1W1N
 
 | | The solution structure of the FATC Domain of the Protein Kinase TOR1 from yeast | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE TOR1 | | Authors: | Dames, S.A, Mulet, J.M, Rathgeb-Szabo, K, Hall, M.N, Grzesiek, S. | | Deposit date: | 2004-06-23 | | Release date: | 2005-03-16 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the FATC domain of the protein kinase target of rapamycin suggests a role for redox-dependent structural and cellular stability.
J. Biol. Chem., 280, 2005
|
|
1W2Q
 
 | |
1W3D
 
 | |
1W4E
 
 | | Peripheral-subunit binding domains from mesophilic, thermophilic, and hyperthermophilic bacteria fold by ultrafast, apparently two-state transitions | | Descriptor: | DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Sato, S, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-Fast Barrier-Limited Folding in the Peripheral Subunit-Binding Domain Family.
J.Mol.Biol., 353, 2005
|
|
1W4F
 
 | | Peripheral-subunit from mesophilic, thermophilic and hyperthermophilic bacteria fold by ultrafast, apparently two-state transitions | | Descriptor: | DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-Fast Barrier-Limited Folding in the Peripheral Subunit-Binding Domain Family.
J.Mol.Biol., 353, 2005
|
|
