1AZ6
 
 | |
1AZE
 
 | | NMR STRUCTURE OF THE COMPLEX BETWEEN THE C32S-Y7V MUTANT OF THE NSH3 DOMAIN OF GRB2 WITH A PEPTIDE FROM SOS, 10 STRUCTURES | | Descriptor: | GRB2, SOS | | Authors: | Vidal, M, Gincel, E, Goudreau, N, Cornille, F, Parker, F, Duchesne, M, Tocque, B, Garbay, C, Roques, B.P. | | Deposit date: | 1997-11-17 | | Release date: | 1999-05-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular and cellular analysis of Grb2 SH3 domain mutants: interaction with Sos and dynamin.
J.Mol.Biol., 290, 1999
|
|
1AZH
 
 | |
1AZJ
 
 | |
1AZK
 
 | |
1B03
 
 | |
1B0S
 
 | | BINDING OF AR-1-144, A TRI-IMIDAZOLE DNA MINOR GROOVE BINDER, TO CCGG SEQUENCE ANALYZED BY NMR SPECTROSCOPY | | Descriptor: | (2-{[4-({4-[(4-FORMYLAMINO-1-METHYL-1H-IMIDAZOLE-2-CARBONYL)-AMINO]-1-METHYL-1H-IMIDAZOLE-2-CARBONYL}-AMINO)-1-METHYL-1 H-IMIDAZOLE-2-CARBONYL]-AMINO}-ETHYL)-DIMETHYL-AMMONIUM, DNA (5'-D(*GP*AP*AP*CP*CP*GP*GP*TP*TP*C)-3') | | Authors: | Yang, X.-L, Kaenzig, C, Lee, M, Wang, A.H.-J. | | Deposit date: | 1998-11-12 | | Release date: | 1999-08-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Binding of AR-1-144, a tri-imidazole DNA minor groove binder, to CCGG sequence analyzed by NMR spectroscopy.
Eur.J.Biochem., 263, 1999
|
|
1B1V
 
 | | NMR STRUCTURE OF PSP1, PLASMATOCYTE-SPREADING PEPTIDE FROM PSEUDOPLUSIA INCLUDENS | | Descriptor: | PROTEIN (PLASMATOCYTE-SPREADING PEPTIDE) | | Authors: | Volkman, B.F, Clark, K.D, Anderson, M.E, Pech, L.L, Markley, J.L, Strand, M.R. | | Deposit date: | 1998-11-23 | | Release date: | 1998-12-02 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of the insect cytokine peptide plasmatocyte-spreading peptide 1 from Pseudoplusia includens.
J.Biol.Chem., 274, 1999
|
|
1B22
 
 | | RAD51 (N-TERMINAL DOMAIN) | | Descriptor: | DNA REPAIR PROTEIN RAD51 | | Authors: | Aihara, H, Ito, Y, Kurumizaka, H, Yokoyama, S, Shibata, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-12-04 | | Release date: | 1999-12-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The N-terminal domain of the human Rad51 protein binds DNA: structure and a DNA binding surface as revealed by NMR.
J.Mol.Biol., 290, 1999
|
|
1B28
 
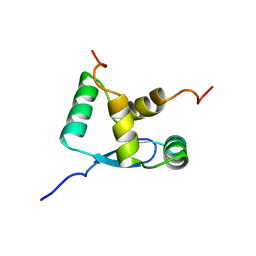 | | ARC REPRESSOR MYL MUTANT FROM SALMONELLA BACTERIOPHAGE P22 | | Descriptor: | PROTEIN (REGULATORY PROTEIN ARC) | | Authors: | Rietveld, A.W.M, Nooren, I.M.A, Sauer, R.T, Kaptein, R, Boelens, R. | | Deposit date: | 1998-12-05 | | Release date: | 1999-11-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure and dynamics of an Arc repressor mutant reveal premelting conformational changes related to DNA binding.
Biochemistry, 38, 1999
|
|
1B2I
 
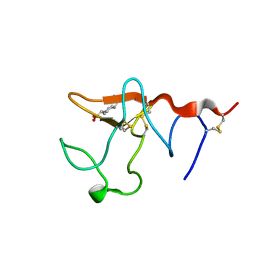 | |
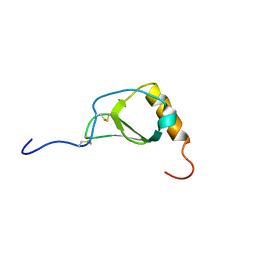
1B2T
 
 | |
1B3C
 
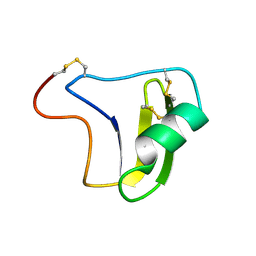 | | SOLUTION STRUCTURE OF A BETA-NEUROTOXIN FROM THE NEW WORLD SCORPION CENTRUROIDES SCULPTURATUS EWING | | Descriptor: | PROTEIN (NEUROTOXIN CSE-I) | | Authors: | Jablonsky, M.J, Jackson, P.L, Trent, J.O, Watt, D.D, Krishna, N.R. | | Deposit date: | 1998-12-08 | | Release date: | 1998-12-16 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a beta-neurotoxin from the New World scorpion Centruroides sculpturatus Ewing.
Biochem.Biophys.Res.Commun., 254, 1999
|
|
1B3P
 
 | | 5'-D(*GP*GP*AP*GP*GP*AP*T)-3' | | Descriptor: | DNA (5'-D(*GP*GP*AP*GP*GP*AP*T)-3') | | Authors: | Kettani, A, Bouaziz, S, Skripkin, E, Majumdar, A, Wang, W, Jones, R.A, Patel, D.J. | | Deposit date: | 1998-12-14 | | Release date: | 1999-08-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Interlocked mismatch-aligned arrowhead DNA motifs.
Structure Fold.Des., 7, 1999
|
|
1B4C
 
 | | SOLUTION STRUCTURE OF RAT APO-S100B USING DIPOLAR COUPLINGS | | Descriptor: | PROTEIN (S-100 PROTEIN, BETA CHAIN) | | Authors: | Weber, D.J, Drohat, A.C, Tjandra, N, Baldisseri, D.M. | | Deposit date: | 1998-12-17 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The use of dipolar couplings for determining the solution structure of rat apo-S100B(betabeta).
Protein Sci., 8, 1999
|
|
1B4M
 
 | | NMR STRUCTURE OF APO CELLULAR RETINOL-BINDING PROTEIN II, 24 STRUCTURES | | Descriptor: | CELLULAR RETINOL-BINDING PROTEIN II | | Authors: | Lu, J, Lin, C.-L, Tang, C, Ponder, J.W, Kao, J.L.F, Cistola, D.P, Li, E. | | Deposit date: | 1998-12-23 | | Release date: | 1999-04-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure and dynamics of rat apo-cellular retinol-binding protein II in solution: comparison with the X-ray structure.
J.Mol.Biol., 286, 1999
|
|
1B4R
 
 | | PKD DOMAIN 1 FROM HUMAN POLYCYSTEIN-1 | | Descriptor: | PROTEIN (PKD1_HUMAN) | | Authors: | Bycroft, M. | | Deposit date: | 1998-12-28 | | Release date: | 1999-01-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of a PKD domain from polycystin-1: implications for polycystic kidney disease.
EMBO J., 18, 1999
|
|
1B4Y
 
 | | STRUCTURE AND MECHANISM OF FORMATION OF THE H-Y5 ISOMER OF AN INTRAMOLECULAR DNA TRIPLE HELIX. | | Descriptor: | DNA (H-Y5 TRIPLE HELIX) | | Authors: | Van Dongen, M.J.P, Doreleijers, J.F, Van Der Marel, G.A, Van Boom, J.H, Hilbers, C.W, Wijmenga, S.S. | | Deposit date: | 1998-12-30 | | Release date: | 1999-09-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and mechanism of formation of the H-y5 isomer of an intramolecular DNA triple helix.
Nat.Struct.Biol., 6, 1999
|
|
1B5K
 
 | | 3,N4-ETHENO-2'-DEOXYCYTIDINE OPPOSITE THYMIDINE IN AN 11-MER DUPLEX, SOLUTION STRUCTURE FROM NMR AND MOLECULAR DYNAMICS | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*CP*EDCP*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*TP*GP*TP*AP*CP*G)-3') | | Authors: | Cullinan, D, Korobka, A, Grollman, A.P, Patel, D.J, Eisenberg, M, De Santos, C.L. | | Deposit date: | 1999-01-07 | | Release date: | 1999-01-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of an oligodeoxynucleotide duplex containing the exocyclic lesion 3,N4-etheno-2'-deoxycytidine opposite thymidine: comparison with the duplex containing deoxyadenosine opposite the adduct.
Biochemistry, 35, 1996
|
|
1B5N
 
 | | NMR STRUCTURE OF PSP1, PLASMATOCYTE-SPREADING PEPTIDE FROM PSEUDOPLUSIA INCLUDENS | | Descriptor: | PROTEIN (PLASMATOCYTE-SPREADING PEPTIDE) | | Authors: | Volkman, B.F, Clark, K.D, Anderson, M.E, Pech, L.L, Markley, J.L, Strand, M.R. | | Deposit date: | 1999-01-07 | | Release date: | 1999-01-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the insect cytokine peptide plasmatocyte-spreading peptide 1 from Pseudoplusia includens.
J.Biol.Chem., 274, 1999
|
|
1B60
 
 | | 3,N4-ETHENO-2'-DEOXYCYTIDINE OPPOSITE CYTIDINE IN AN 11-MER DUPLEX, SOLUTION STRUCTURE FROM NMR AND MOLECULAR DYNAMICS | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*CP*(EDC)P*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*GP*TP*AP*CP*G)-3') | | Authors: | Cullinan, D, Johnson, F, De Los Santos, C. | | Deposit date: | 1999-01-20 | | Release date: | 2000-02-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an 11-mer duplex containing the 3, N(4)-ethenocytosine adduct opposite 2'-deoxycytidine: implications for the recognition of exocyclic lesions by DNA glycosylases.
J.Mol.Biol., 296, 2000
|
|
1B64
 
 | | SOLUTION STRUCTURE OF THE GUANINE NUCLEOTIDE EXCHANGE FACTOR DOMAIN FROM HUMAN ELONGATION FACTOR-ONE BETA, NMR, 20 STRUCTURES | | Descriptor: | ELONGATION FACTOR 1-BETA | | Authors: | Perez, J.M.J, Siegal, G, Kriek, J, Hard, K, Dijk, J, Canters, G.W, Moller, W. | | Deposit date: | 1999-01-20 | | Release date: | 1999-05-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the guanine nucleotide exchange domain of human elongation factor 1beta reveals a striking resemblance to that of EF-Ts from Escherichia coli.
Structure Fold.Des., 7, 1999
|
|
1B6X
 
 | | 3,N4-ETHENO-2'-DEOXYCYTIDINE OPPOSITE GUANINE IN AN 11-MER DUPLEX, SOLUTION STRUCTURE FROM NMR AND MOLECULAR DYNAMICS, 4 STRUCTURES | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*(EDC)P*CP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*GP*GP*GP*TP*AP*CP*G)-3' | | Authors: | Cullinan, D, Johnson, F, Grollman, A.P, Eisenberg, M, De Los Santos, C. | | Deposit date: | 1999-01-19 | | Release date: | 1999-01-27 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA duplex containing the exocyclic lesion 3,N4-etheno-2'-deoxycytidine opposite 2'-deoxyguanosine.
Biochemistry, 36, 1997
|
|
1B6Y
 
 | | 3,N4-ETHENO-2'-DEOXYCYTIDINE OPPOSITE ADENINE IN AN 11-MER DUPLEX, SOLUTION STRUCTURE FROM NMR AND MOLECULAR DYNAMICS, 2 STRUCTURES | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*(EDC)P*CP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*GP*AP*GP*TP*AP*CP*G)-3' | | Authors: | Korobka, A, Cullinan, D, Cosman, M, Grollman, A.P, Patel, D.J, Eisenberg, M, De Los Santos, C. | | Deposit date: | 1999-01-19 | | Release date: | 1999-01-27 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an oligodeoxynucleotide duplex containing the exocyclic lesion 3,N4-etheno-2'-deoxycytidine opposite 2'-deoxyadenosine, determined by NMR spectroscopy and restrained molecular dynamics.
Biochemistry, 35, 1996
|
|
1B75
 
 | |
