1EF4
 
 | | SOLUTION STRUCTURE OF THE ESSENTIAL RNA POLYMERASE SUBUNIT RPB10 FROM METHANOBACTERIUM THERMOAUTOTROPHICUM | | Descriptor: | DNA-DIRECTED RNA POLYMERASE, ZINC ION | | Authors: | Mackereth, C.D, Arrowsmith, C.H, Edwards, A.M, Mcintosh, L.P, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-02-07 | | Release date: | 2000-06-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Zinc-bundle structure of the essential RNA polymerase subunit RPB10 from Methanobacterium thermoautotrophicum.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EF5
 
 | | SOLUTION STRUCTURE OF THE RAS-BINDING DOMAIN OF RGL | | Descriptor: | RGL | | Authors: | Kigawa, T, Endo, M, Ito, Y, Shirouzu, M, Kikuchi, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-02-07 | | Release date: | 2000-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Ras-binding domain of RGL.
FEBS Lett., 441, 1998
|
|
1EFE
 
 | | AN ACTIVE MINI-PROINSULIN, M2PI | | Descriptor: | MINI-PROINSULIN | | Authors: | Cho, Y, Chang, S.G, Choi, K.D, Shin, H, Ahn, B, Kim, K.S. | | Deposit date: | 2000-02-08 | | Release date: | 2000-03-17 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of an Active Mini-Proinsulin, M2PI: Inter-chain Flexibility is Crucial for Insulin Activity
J.Biochem.Mol.Biol., 33, 2000
|
|
1EFS
 
 | | CONFORMATION OF A DNA-RNA HYBRID | | Descriptor: | DNA (5'-D(*GP*AP*GP*AP*GP*GP*AP*AP*GP*AP*GP*AP*A)-3'), RNA (5'-R(*UP*UP*CP*UP*CP*UP*UP*CP*CP*UP*CP*UP*C)-3') | | Authors: | Larue, V, Hantz, E. | | Deposit date: | 2000-02-10 | | Release date: | 2000-03-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of an RNA--DNA hybrid duplex containing a pyrimidine RNA strand and a purine DNA strand.
Int.J.Biol.Macromol., 28, 2001
|
|
1EGF
 
 | |
1EGO
 
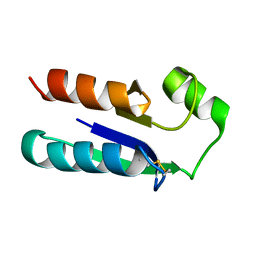 | | NMR STRUCTURE OF OXIDIZED ESCHERICHIA COLI GLUTAREDOXIN: COMPARISON WITH REDUCED E. COLI GLUTAREDOXIN AND FUNCTIONALLY RELATED PROTEINS | | Descriptor: | GLUTAREDOXIN | | Authors: | Xia, T.-H, Bushweller, J.H, Sodano, P, Billeter, M, Bjornberg, O, Holmgren, A, Wuthrich, K. | | Deposit date: | 1991-10-08 | | Release date: | 1993-10-31 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of oxidized Escherichia coli glutaredoxin: comparison with reduced E. coli glutaredoxin and functionally related proteins.
Protein Sci., 1, 1992
|
|
1EGR
 
 | | SEQUENCE-SPECIFIC 1H N.M.R. ASSIGNMENTS AND DETERMINATION OF THE THREE-DIMENSIONAL STRUCTURE OF REDUCED ESCHERICHIA COLI GLUTAREDOXIN | | Descriptor: | GLUTAREDOXIN | | Authors: | Sodano, P, Xia, T.-H, Bushweller, J.H, Bjornberg, O, Holmgren, A, Billeter, M, Wuthrich, K. | | Deposit date: | 1991-10-08 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Sequence-specific 1H n.m.r. assignments and determination of the three-dimensional structure of reduced Escherichia coli glutaredoxin.
J.Mol.Biol., 221, 1991
|
|
1EGS
 
 | |
1EGX
 
 | | SOLUTION STRUCTURE OF THE ENA-VASP HOMOLOGY 1 (EVH1) DOMAIN OF HUMAN VASODILATOR-STIMULATED PHOSPHOPROTEIN (VASP) | | Descriptor: | VASODILATOR-STIMULATED PHOSPHOPROTEIN | | Authors: | Ball, L, Kuhne, R, Hoffmann, B, Hafner, A, Schmieder, P, Volkmer-Engert, R, Hof, M, Wahl, M, Schneider-Mergener, J, Walter, U, Oschkinat, H, Jarchau, T. | | Deposit date: | 2000-02-17 | | Release date: | 2000-09-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Dual epitope recognition by the VASP EVH1 domain modulates polyproline ligand specificity and binding affinity.
EMBO J., 19, 2000
|
|
1EH2
 
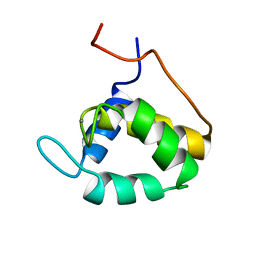 | | STRUCTURE OF THE SECOND EPS15 HOMOLOGY DOMAIN OF HUMAN EPS15, NMR, 20 STRUCTURES | | Descriptor: | CALCIUM ION, EPS15 | | Authors: | De Beer, T, Carter, R.E, Lobel-Rice, K.E, Sorkin, A, Overduin, M. | | Deposit date: | 1998-07-10 | | Release date: | 1999-07-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and Asn-Pro-Phe binding pocket of the Eps15 homology domain.
Science, 281, 1998
|
|
1EHJ
 
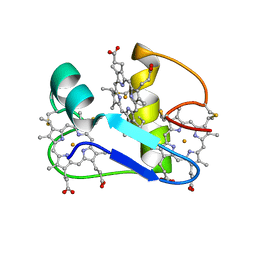 | | A PROTON-NMR INVESTIGATION OF THE FULLY REDUCED CYTOCHROME C7 FROM DESULFUROMONAS ACETOXIDANS | | Descriptor: | CYTOCHROME C7, HEME C | | Authors: | Assfalg, M, Banci, L, Bertini, I, Bruschi, M, Giudici-Orticoni, M.T. | | Deposit date: | 2000-02-21 | | Release date: | 2000-05-10 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | A proton-NMR investigation of the fully reduced cytochrome c7 from Desulfuromonas acetoxidans. Comparison between the reduced and the oxidized forms.
Eur.J.Biochem., 266, 1999
|
|
1EHT
 
 | |
1EHX
 
 | | NMR SOLUTION STRUCTURE OF THE LAST UNKNOWN MODULE OF THE CELLULOSOMAL SCAFFOLDIN PROTEIN CIPC OF CLOSTRIDUM CELLULOLYTICUM | | Descriptor: | SCAFFOLDIN PROTEIN | | Authors: | Mosbah, A, Belaich, A, Bornet, O, Belaich, J.P, Henrissat, B, Darbon, H. | | Deposit date: | 2000-02-23 | | Release date: | 2000-11-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the module X2 1 of unknown function of the cellulosomal scaffolding protein CipC of Clostridium cellulolyticum.
J.Mol.Biol., 304, 2000
|
|
1EIG
 
 | |
1EIH
 
 | |
1EII
 
 | | NMR STRUCTURE OF HOLO CELLULAR RETINOL-BINDING PROTEIN II | | Descriptor: | CELLULAR RETINOL-BINDING PROTEIN II, RETINOL | | Authors: | Lu, J, Lin, C.L, Tang, C, Ponder, J.W, Kao, J.L, Cistola, D.P, Li, E. | | Deposit date: | 2000-02-25 | | Release date: | 2000-08-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Binding of retinol induces changes in rat cellular retinol-binding protein II conformation and backbone dynamics.
J.Mol.Biol., 300, 2000
|
|
1EIO
 
 | | ILEAL LIPID BINDING PROTEIN IN COMPLEX WITH GLYCOCHOLATE | | Descriptor: | GLYCOCHOLIC ACID, ILEAL LIPID BINDING PROTEIN | | Authors: | Luecke, C, Zhang, F, Hamilton, J.A, Sacchettini, J.C, Rueterjans, H. | | Deposit date: | 2000-02-27 | | Release date: | 2000-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ileal lipid binding protein in complex with glycocholate.
Eur.J.Biochem., 267, 2000
|
|
1EIT
 
 | | NMR STUDY OF MU-AGATOXIN | | Descriptor: | MU-AGATOXIN-I | | Authors: | Omecinsky, D.O, Reily, M.D. | | Deposit date: | 1995-12-14 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure analysis of mu-agatoxins: further evidence for common motifs among neurotoxins with diverse ion channel specificities.
Biochemistry, 35, 1996
|
|
1EIW
 
 | |
1EJP
 
 | | SOLUTION STRUCTURE OF THE SYNDECAN-4 WHOLE CYTOPLASMIC DOMAIN | | Descriptor: | SYNDECAN-4 | | Authors: | Lee, D, Oh, E.S, Woods, A, Couchman, J.R, Lee, W. | | Deposit date: | 2000-03-03 | | Release date: | 2001-09-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the dimeric cytoplasmic domain of syndecan-4.
Biochemistry, 40, 2001
|
|
1EJQ
 
 | | SOLUTION STRUCTURE OF THE SYNDECAN-4 WHOLE CYTOPLASMIC DOMAIN IN THE PRESENCE OF PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE | | Descriptor: | SYNDECAN-4 | | Authors: | Shin, J, Oh, E.S, Lee, D, Couchman, J.R, Lee, W. | | Deposit date: | 2000-03-04 | | Release date: | 2001-03-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | SOLUTION STRUCTURE OF THE SYNDECAN-4 WHOLE CYTOPLASMIC DOMAIN IN THE PRESENCE OF PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE
To be Published
|
|
1EKA
 
 | | NMR AND MOLECULAR MODELING REVEAL THAT DIFFERENT HYDROGEN BONDING PATTERNS ARE POSSIBLE FOR GU PAIRS: ONE HYDROGEN BOND FOR EACH GU PAIR IN R(GGCGUGCC)2 AND TWO FOR EACH GU PAIR IN R(GAGUGCUC)2 | | Descriptor: | RNA (5'-R(*GP*AP*GP*UP*GP*CP*UP*C)-3') | | Authors: | Chen, X, McDowell, J.A, Kierzek, R, Krugh, T.R, Turner, D.H. | | Deposit date: | 2000-03-07 | | Release date: | 2000-11-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance spectroscopy and molecular modeling reveal that different hydrogen bonding patterns are possible for G.U pairs: one hydrogen bond for each G.U pair in r(GGCGUGCC)(2) and two for each G.U pair in r(GAGUGCUC)(2).
Biochemistry, 39, 2000
|
|
1EKD
 
 | | NMR AND MOLECULAR MODELING REVEAL THAT DIFFERENT HYDROGEN BONDING PATTERNS ARE POSSIBLE FOR GU PAIRS: ONE HYDROGEN BOND FOR EACH GU PAIR IN R(GGCGUGCC)2 AND TWO FOR EACH GU PAIR IN R(GAGUGCUC)2 | | Descriptor: | RNA (5'-R(*GP*GP*CP*GP*UP*GP*CP*C)-3') | | Authors: | Chen, X, McDowell, J.A, Kierzek, R, Krugh, T.R, Turner, D.H. | | Deposit date: | 2000-03-07 | | Release date: | 2000-11-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance spectroscopy and molecular modeling reveal that different hydrogen bonding patterns are possible for G.U pairs: one hydrogen bond for each G.U pair in r(GGCGUGCC)(2) and two for each G.U pair in r(GAGUGCUC)(2).
Biochemistry, 39, 2000
|
|
1EKH
 
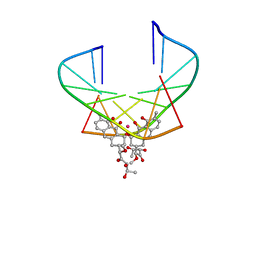 | | NMR STRUCTURE OF D(TTGGCCAA)2 BOUND TO CHROMOMYCIN-A3 AND COBALT | | Descriptor: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-4-O-acetyl-2,6-dideoxy-beta-D-galactopyranose, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-beta-D-Olivopyranose-(1-3)-beta-D-Olivopyranose, ... | | Authors: | Gochin, M. | | Deposit date: | 2000-03-08 | | Release date: | 2000-03-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A high-resolution structure of a DNA-chromomycin-Co(II) complex determined from pseudocontact shifts in nuclear magnetic resonance.
Structure Fold.Des., 8, 2000
|
|
1EL2
 
 | |
