7MUC
 
 | | Legionella pneumophila Dot/Icm T4SS C1 Reconstruction | | Descriptor: | DUF2807 domain-containing protein, DotC, DotD, ... | | Authors: | Sheedlo, M.J, Durie, C.L, Swanson, M, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2021-05-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM reveals new species-specific proteins and symmetry elements in the Legionella pneumophila Dot/Icm T4SS.
Elife, 10, 2021
|
|
7MUD
 
 | | Legionella pneumophila Dot/Icm T4SS OMC | | Descriptor: | DUF2807 domain-containing protein, DotC, DotD, ... | | Authors: | Sheedlo, M.J, Durie, C.L, Swanson, M, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2021-05-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM reveals new species-specific proteins and symmetry elements in the Legionella pneumophila Dot/Icm T4SS.
Elife, 10, 2021
|
|
7MUE
 
 | | Legionella pneumophila Dot/Icm T4SS PR | | Descriptor: | DotF, IcmE protein, Type IV secretion protein IcmK, ... | | Authors: | Sheedlo, M.J, Durie, C.L, Swanson, M, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2021-05-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM reveals new species-specific proteins and symmetry elements in the Legionella pneumophila Dot/Icm T4SS.
Elife, 10, 2021
|
|
7MUQ
 
 | | Reconstruction of the Legionella pneumophila Dot/Icm T4SS 3DVA Map 1 | | Descriptor: | DUF2807 domain-containing protein, DotC, DotD, ... | | Authors: | Sheedlo, M.J, Durie, C.L, Swanson, M, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2021-05-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM reveals new species-specific proteins and symmetry elements in the Legionella pneumophila Dot/Icm T4SS.
Elife, 10, 2021
|
|
7MUS
 
 | | Reconstruction of the Legionella pneumophila Dot/Icm T4SS 3DVA Map 2 | | Descriptor: | DUF2807 domain-containing protein, DotC, DotD, ... | | Authors: | Sheedlo, M.J, Durie, C.L, Swanson, M, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2021-05-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM reveals new species-specific proteins and symmetry elements in the Legionella pneumophila Dot/Icm T4SS.
Elife, 10, 2021
|
|
7MUV
 
 | | Reconstruction of the Legionella pneumophila Dot/Icm T4SS 3DVA Map 3 | | Descriptor: | DUF2807 domain-containing protein, DotC, DotD, ... | | Authors: | Sheedlo, M.J, Durie, C.L, Swanson, M, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2021-05-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM reveals new species-specific proteins and symmetry elements in the Legionella pneumophila Dot/Icm T4SS.
Elife, 10, 2021
|
|
7MUW
 
 | | Reconstruction of the Legionella pneumophila Dot/Icm T4SS 3DVA Map 4 | | Descriptor: | DUF2807 domain-containing protein, DotC, DotD, ... | | Authors: | Sheedlo, M.J, Durie, C.L, Swanson, M, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2021-05-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM reveals new species-specific proteins and symmetry elements in the Legionella pneumophila Dot/Icm T4SS.
Elife, 10, 2021
|
|
7MUY
 
 | | Reconstruction of the Legionella pneumophila Dot/Icm T4SS 3DVA Map 5 | | Descriptor: | DUF2807 domain-containing protein, DotC, DotD, ... | | Authors: | Sheedlo, M.J, Durie, C.L, Swanson, M, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2021-05-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM reveals new species-specific proteins and symmetry elements in the Legionella pneumophila Dot/Icm T4SS.
Elife, 10, 2021
|
|
7MX6
 
 | | Leishmania major dihydroorotate dehydrogenase in complex with [4-(1H-pyrrol-1-yl)phenyl]methanol | | Descriptor: | 4-(1H-pyrrol-1-yl)aniline, Dihydroorotate dehydrogenase (fumarate), FLAVIN MONONUCLEOTIDE, ... | | Authors: | Pinheiro, M.P, Hunter, W.N, Cardoso, I.A, Nonato, M.C. | | Deposit date: | 2021-05-18 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Leishmania major dihydroorotate dehydrogenase in complex with [4-(1H-pyrrol-1-yl)phenyl]methanol
To be Published
|
|
7MXZ
 
 | | Sy-CrtE apo structure | | Descriptor: | CHLORIDE ION, Geranylgeranyl pyrophosphate synthase, MAGNESIUM ION | | Authors: | Peat, T.S, Newman, J. | | Deposit date: | 2021-05-19 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Molecular characterization of cyanobacterial short-chain prenyltransferases and discovery of a novel GGPP phosphatase.
Febs J., 289, 2022
|
|
7MY1
 
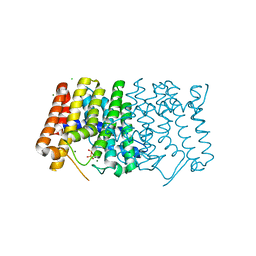 | | Sy-CrtE structure with IPP, N-term His-tag | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, CHLORIDE ION, Geranylgeranyl pyrophosphate synthase, ... | | Authors: | Peat, T.S, Newman, J. | | Deposit date: | 2021-05-19 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Molecular characterization of cyanobacterial short-chain prenyltransferases and discovery of a novel GGPP phosphatase.
Febs J., 289, 2022
|
|
7MY7
 
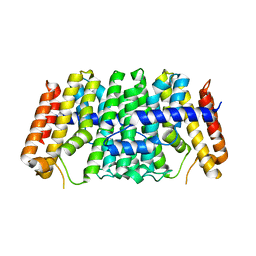 | | Se-CrtE N-term His-tag structure | | Descriptor: | Farnesyl-diphosphate synthase | | Authors: | Peat, T.S, Newman, J. | | Deposit date: | 2021-05-20 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Molecular characterization of cyanobacterial short-chain prenyltransferases and discovery of a novel GGPP phosphatase.
Febs J., 289, 2022
|
|
7MYD
 
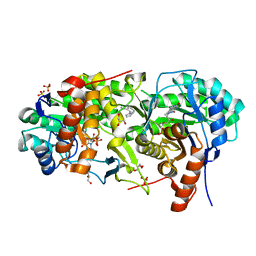 | | Leishmania major dihydroorotate dehydrogenase in complex with 5-amino-2-(1H-pyrrol-1-yl)benzonitrile | | Descriptor: | 5-azanyl-2-pyrrol-1-yl-benzenecarbonitrile, Dihydroorotate dehydrogenase (fumarate), FLAVIN MONONUCLEOTIDE, ... | | Authors: | Pinheiro, M.P, Cardoso, I.A, Hunter, W.N, Nonato, M.C. | | Deposit date: | 2021-05-20 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Leishmania major dihydroorotate dehydrogenase in complex with 5-amino-2-(1H-pyrrol-1-yl)benzonitrile
To Be Published
|
|
7N1H
 
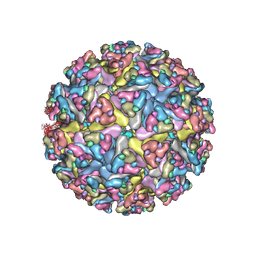 | | CryoEM structure of Venezuelan equine encephalitis virus VLP in complex with the LDLRAD3 receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Capsid, ... | | Authors: | Basore, K, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-27 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus in complex with the LDLRAD3 receptor.
Nature, 598, 2021
|
|
7N1I
 
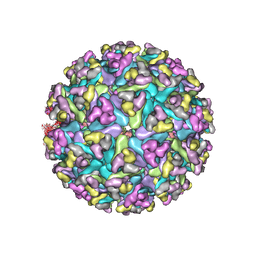 | | CryoEM structure of Venezuelan equine encephalitis virus VLP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid, E1 envelope glycoprotein, ... | | Authors: | Basore, K, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-27 | | Release date: | 2021-10-13 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus in complex with the LDLRAD3 receptor.
Nature, 598, 2021
|
|
7N1P
 
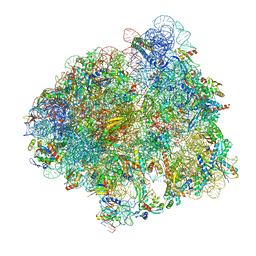 | | Elongating 70S ribosome complex in a classical pre-translocation (PRE-C) conformation | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, S.K, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-14 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
7N2C
 
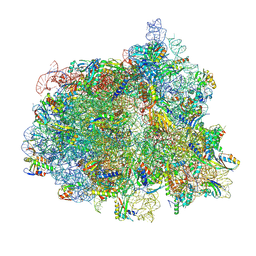 | | Elongating 70S ribosome complex in a fusidic acid-stalled intermediate state of translocation bound to EF-G(GDP) (INT2) | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-14 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
7N2U
 
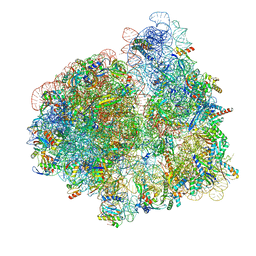 | | Elongating 70S ribosome complex in a hybrid-H1 pre-translocation (PRE-H1) conformation | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-29 | | Release date: | 2021-07-14 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
7N2V
 
 | | Elongating 70S ribosome complex in a spectinomycin-stalled intermediate state of translocation bound to EF-G in an active, GTP conformation (INT1) | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-29 | | Release date: | 2021-07-14 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
7N30
 
 | | Elongating 70S ribosome complex in a hybrid-H2* pre-translocation (PRE-H2*) conformation | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-30 | | Release date: | 2021-07-14 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
7N31
 
 | | Elongating 70S ribosome complex in a post-translocation (POST) conformation | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-31 | | Release date: | 2021-07-14 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
7N3M
 
 | | Co-complex CYP46A1 with 0431 (compound 17) | | Descriptor: | Cholesterol 24-hydroxylase, N,N-dimethyl-1-[4-(4-methyl-1H-pyrazol-1-yl)pyridin-3-yl]piperidine-4-carboxamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lane, W, Yano, J. | | Deposit date: | 2021-06-01 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Discovery of Novel 3-Piperidinyl Pyridine Derivatives as Highly Potent and Selective Cholesterol 24-Hydroxylase (CH24H) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7N50
 
 | |
7N51
 
 | |
7N52
 
 | |







