[English] 日本語
 Yorodumi
Yorodumi- PDB-6g8h: Flavonoid-responsive Regulator FrrA in complex with (R,S)-Naringenin -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6g8h | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Flavonoid-responsive Regulator FrrA in complex with (R,S)-Naringenin | |||||||||
 Components Components | Transcriptional regulatory protein | |||||||||
 Keywords Keywords |  TRANSCRIPTION / TRANSCRIPTION /  Flavonoids / Flavonoids /  repressor / TetR-family repressor / TetR-family | |||||||||
| Function / homology |  Function and homology information Function and homology informationtranscription cis-regulatory region binding / DNA-binding transcription factor activity / regulation of DNA-templated transcription Similarity search - Function | |||||||||
| Biological species |   Bradyrhizobium diazoefficiens (bacteria) Bradyrhizobium diazoefficiens (bacteria) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.6 Å MOLECULAR REPLACEMENT / Resolution: 2.6 Å | |||||||||
 Authors Authors | Werner, N. / Hoppen, J. / Palm, G. / Werten, S. / Goettfert, M. / Hinrichs, W. | |||||||||
 Citation Citation |  Journal: Febs J. / Year: 2021 Journal: Febs J. / Year: 2021Title: The induction mechanism of the flavonoid-responsive regulator FrrA. Authors: Werner, N. / Werten, S. / Hoppen, J. / Palm, G.J. / Gottfert, M. / Hinrichs, W. #1: Journal: Journal of Bacteriology / Year: 2012 Title: Characterization of the Flavonoid-Responsive Regulator FrrA and Its Binding Sites Authors: Wenzel, M. / Lang, K. / Gottfert, M. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6g8h.cif.gz 6g8h.cif.gz | 297.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6g8h.ent.gz pdb6g8h.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  6g8h.json.gz 6g8h.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/g8/6g8h https://data.pdbj.org/pub/pdb/validation_reports/g8/6g8h ftp://data.pdbj.org/pub/pdb/validation_reports/g8/6g8h ftp://data.pdbj.org/pub/pdb/validation_reports/g8/6g8h | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6g87SC  6g8gC S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data | |
| Other databases |
|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Unit cell |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments:
NCS ensembles :
|
- Components
Components
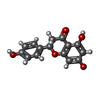
| #1: Protein | Mass: 21903.803 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Details: The sequence represents the crystallized polypeptide Source: (gene. exp.)   Bradyrhizobium diazoefficiens (strain JCM 10833 / BCRC 13528 / IAM 13628 / NBRC 14792 / USDA 110) (bacteria) Bradyrhizobium diazoefficiens (strain JCM 10833 / BCRC 13528 / IAM 13628 / NBRC 14792 / USDA 110) (bacteria)Strain: JCM 10833 / BCRC 13528 / IAM 13628 / NBRC 14792 / USDA 110 Gene: blr4322 / Production host:   Escherichia coli BL21(DE3) (bacteria) / Variant (production host): Gold pLysS AG / References: UniProt: Q89M71 Escherichia coli BL21(DE3) (bacteria) / Variant (production host): Gold pLysS AG / References: UniProt: Q89M71#2: Chemical | ChemComp-NHE /  CHES (buffer) CHES (buffer)#3: Chemical | ChemComp-CWE / | #4: Chemical | ChemComp-NAR / |  Naringenin NaringeninHas ligand of interest | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.88 Å3/Da / Density % sol: 57.24 % |
|---|---|
Crystal grow | Temperature: 291 K / Method: vapor diffusion, hanging drop Details: 6mg/ml FrrA in 20 mM Na2HPO4, 50 mM imidazole, pH 7.5, 50 mM NaCl with equimolar naringenin in 70% Ethanol. Precipitant 16,5 % PEG 8000; 0.1 M CHES, pH 9.5. PH range: 7.5 - 9.5 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  PETRA III, EMBL c/o DESY PETRA III, EMBL c/o DESY  / Beamline: P13 (MX1) / Wavelength: 0.9763 Å / Beamline: P13 (MX1) / Wavelength: 0.9763 Å |
| Detector | Type: DECTRIS PILATUS3 S 6M / Detector: PIXEL / Date: Oct 27, 2015 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.9763 Å / Relative weight: 1 : 0.9763 Å / Relative weight: 1 |
| Reflection | Resolution: 2.312→118.71 Å / Num. obs: 67523 / % possible obs: 95.2 % / Redundancy: 3.35 % / Biso Wilson estimate: 81.2 Å2 / CC1/2: 0.998 / Rrim(I) all: 0.094 / Net I/σ(I): 9.74 |
| Reflection shell | Resolution: 2.312→2.54 Å / Redundancy: 2.76 % / Mean I/σ(I) obs: 0.54 / Num. unique obs: 8294 / CC1/2: 0.2 / Rrim(I) all: 1.76 / % possible all: 72.2 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 6G87 Resolution: 2.6→118.709 Å / Cor.coef. Fo:Fc: 0.967 / Cor.coef. Fo:Fc free: 0.953 / SU B: 27.121 / SU ML: 0.242 / Cross valid method: FREE R-VALUE / ESU R: 0.444 / ESU R Free: 0.254 Details: Hydrogens have been added in their riding positions; TLS refinement
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK BULK SOLVENT | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 75.586 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.6→118.709 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj