[English] 日本語
 Yorodumi
Yorodumi- PDB-1irs: IRS-1 PTB DOMAIN COMPLEXED WITH A IL-4 RECEPTOR PHOSPHOPEPTIDE, N... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1irs | ||||||
|---|---|---|---|---|---|---|---|
| Title | IRS-1 PTB DOMAIN COMPLEXED WITH A IL-4 RECEPTOR PHOSPHOPEPTIDE, NMR, MINIMIZED AVERAGE STRUCTURE | ||||||
 Components Components |
| ||||||
 Keywords Keywords | COMPLEX (SIGNAL TRANSDUCTION/PEPTIDE) / PHOSPHOTYROSINE BINDING DOMAIN (PTB) /  COMPLEX / COMPLEX /  SIGNAL TRANSDUCTION / COMPLEX (SIGNAL TRANSDUCTION-PEPTIDE) / COMPLEX (SIGNAL TRANSDUCTION-PEPTIDE) complex SIGNAL TRANSDUCTION / COMPLEX (SIGNAL TRANSDUCTION-PEPTIDE) / COMPLEX (SIGNAL TRANSDUCTION-PEPTIDE) complex | ||||||
| Function / homology |  Function and homology information Function and homology information interleukin-4 receptor activity / production of molecular mediator involved in inflammatory response / negative regulation of T-helper 1 cell differentiation / T-helper 1 cell differentiation / positive regulation of T-helper 2 cell differentiation / IRS-related events triggered by IGF1R / interleukin-4-mediated signaling pathway / IRS-mediated signalling / positive regulation of fatty acid beta-oxidation / positive regulation of mast cell degranulation ... interleukin-4 receptor activity / production of molecular mediator involved in inflammatory response / negative regulation of T-helper 1 cell differentiation / T-helper 1 cell differentiation / positive regulation of T-helper 2 cell differentiation / IRS-related events triggered by IGF1R / interleukin-4-mediated signaling pathway / IRS-mediated signalling / positive regulation of fatty acid beta-oxidation / positive regulation of mast cell degranulation ... interleukin-4 receptor activity / production of molecular mediator involved in inflammatory response / negative regulation of T-helper 1 cell differentiation / T-helper 1 cell differentiation / positive regulation of T-helper 2 cell differentiation / IRS-related events triggered by IGF1R / interleukin-4-mediated signaling pathway / IRS-mediated signalling / positive regulation of fatty acid beta-oxidation / positive regulation of mast cell degranulation / interleukin-4 receptor activity / production of molecular mediator involved in inflammatory response / negative regulation of T-helper 1 cell differentiation / T-helper 1 cell differentiation / positive regulation of T-helper 2 cell differentiation / IRS-related events triggered by IGF1R / interleukin-4-mediated signaling pathway / IRS-mediated signalling / positive regulation of fatty acid beta-oxidation / positive regulation of mast cell degranulation /  insulin receptor complex / positive regulation of glucose metabolic process / positive regulation of macrophage activation / Activated NTRK3 signals through PI3K / insulin receptor complex / positive regulation of glucose metabolic process / positive regulation of macrophage activation / Activated NTRK3 signals through PI3K /  transmembrane receptor protein tyrosine kinase adaptor activity / T-helper 2 cell differentiation / Signaling by Leptin / transmembrane receptor protein tyrosine kinase adaptor activity / T-helper 2 cell differentiation / Signaling by Leptin /  cytokine receptor activity / PI3K/AKT activation / cellular response to fatty acid / Signaling by ALK / positive regulation of myoblast fusion / IRS activation / defense response to protozoan / immunoglobulin mediated immune response / positive regulation of immunoglobulin production / centriolar satellite / Signaling by ALK fusions and activated point mutants / SOS-mediated signalling / PI3K Cascade / negative regulation of insulin secretion / positive regulation of glycogen biosynthetic process / Signal attenuation / cytokine receptor activity / PI3K/AKT activation / cellular response to fatty acid / Signaling by ALK / positive regulation of myoblast fusion / IRS activation / defense response to protozoan / immunoglobulin mediated immune response / positive regulation of immunoglobulin production / centriolar satellite / Signaling by ALK fusions and activated point mutants / SOS-mediated signalling / PI3K Cascade / negative regulation of insulin secretion / positive regulation of glycogen biosynthetic process / Signal attenuation /  phosphatidylinositol 3-kinase binding / Growth hormone receptor signaling / positive regulation of insulin receptor signaling pathway / positive regulation of chemokine production / negative regulation of insulin receptor signaling pathway / phosphatidylinositol 3-kinase binding / Growth hormone receptor signaling / positive regulation of insulin receptor signaling pathway / positive regulation of chemokine production / negative regulation of insulin receptor signaling pathway /  insulin-like growth factor receptor binding / Interleukin-7 signaling / insulin-like growth factor receptor binding / Interleukin-7 signaling /  SH2 domain binding / phosphotyrosine residue binding / SH2 domain binding / phosphotyrosine residue binding /  protein kinase C binding / insulin-like growth factor receptor signaling pathway / phosphatidylinositol 3-kinase/protein kinase B signal transduction / protein kinase C binding / insulin-like growth factor receptor signaling pathway / phosphatidylinositol 3-kinase/protein kinase B signal transduction /  caveola / positive regulation of glucose import / response to insulin / caveola / positive regulation of glucose import / response to insulin /  insulin receptor binding / cytokine-mediated signaling pathway / cellular response to insulin stimulus / Constitutive Signaling by Aberrant PI3K in Cancer / signaling receptor complex adaptor activity / insulin receptor binding / cytokine-mediated signaling pathway / cellular response to insulin stimulus / Constitutive Signaling by Aberrant PI3K in Cancer / signaling receptor complex adaptor activity /  glucose homeostasis / PIP3 activates AKT signaling / insulin receptor signaling pathway / positive regulation of cold-induced thermogenesis / PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling / RAF/MAP kinase cascade / Interleukin-4 and Interleukin-13 signaling / positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / glucose homeostasis / PIP3 activates AKT signaling / insulin receptor signaling pathway / positive regulation of cold-induced thermogenesis / PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling / RAF/MAP kinase cascade / Interleukin-4 and Interleukin-13 signaling / positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction /  receptor complex / receptor complex /  immune response / external side of plasma membrane / intracellular membrane-bounded organelle / positive regulation of cell population proliferation / immune response / external side of plasma membrane / intracellular membrane-bounded organelle / positive regulation of cell population proliferation /  signal transduction / extracellular region / signal transduction / extracellular region /  nucleoplasm / nucleoplasm /  nucleus / nucleus /  plasma membrane / plasma membrane /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  SOLUTION NMR SOLUTION NMR | ||||||
 Authors Authors | Zhou, M.-M. / Huang, B. / Olejniczak, E.T. / Meadows, R.P. / Shuker, S.B. / Miyazaki, M. / Trub, T. / Shoelson, S.E. / Feisk, S.W. | ||||||
 Citation Citation |  Journal: Nat.Struct.Biol. / Year: 1996 Journal: Nat.Struct.Biol. / Year: 1996Title: Structural basis for IL-4 receptor phosphopeptide recognition by the IRS-1 PTB domain. Authors: Zhou, M.M. / Huang, B. / Olejniczak, E.T. / Meadows, R.P. / Shuker, S.B. / Miyazaki, M. / Trub, T. / Shoelson, S.E. / Fesik, S.W. #1:  Journal: J.Biol.Chem. / Year: 1995 Journal: J.Biol.Chem. / Year: 1995Title: Ptb Domains of Irs-1 and Shc Have Distinct But Overlapping Binding Specificities Authors: Wolf, G. / Trub, T. / Ottinger, E. / Groninga, L. / Lynch, A. / White, M.F. / Miyazaki, M. / Lee, J. / Shoelson, S.E. #2:  Journal: Nature / Year: 1995 Journal: Nature / Year: 1995Title: Structure and Ligand Recognition of the Phosphotyrosine Binding Domain of Shc Authors: Zhou, M.M. / Ravichandran, K.S. / Olejniczak, E.F. / Petros, A.M. / Meadows, R.P. / Sattler, M. / Harlan, J.E. / Wade, W.S. / Burakoff, S.J. / Fesik, S.W. #3:  Journal: Mol.Cell.Biol. / Year: 1994 Journal: Mol.Cell.Biol. / Year: 1994Title: Characterization of an Interaction between Insulin Receptor Substrate 1 and the Insulin Receptor by Using the Two-Hybrid System Authors: O'Neill, T.J. / Craparo, A. / Gustafson, T.A. | ||||||
| History |
|
- Structure visualization
Structure visualization
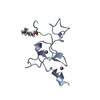
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1irs.cif.gz 1irs.cif.gz | 52.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1irs.ent.gz pdb1irs.ent.gz | 40.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1irs.json.gz 1irs.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ir/1irs https://data.pdbj.org/pub/pdb/validation_reports/ir/1irs ftp://data.pdbj.org/pub/pdb/validation_reports/ir/1irs ftp://data.pdbj.org/pub/pdb/validation_reports/ir/1irs | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein |  IRS1 / INSULIN RECEPTOR SUBSTRATE 1 IRS1 / INSULIN RECEPTOR SUBSTRATE 1Mass: 12648.737 Da / Num. of mol.: 1 / Fragment: PTB DOMAIN Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Strain: BL21 (DE2) PLYSS / Cell line: BL21 / Gene: PTB DOMAIN OF IRS-1 / Organ: SKELETAL Homo sapiens (human) / Strain: BL21 (DE2) PLYSS / Cell line: BL21 / Gene: PTB DOMAIN OF IRS-1 / Organ: SKELETAL Skeleton / Plasmid: PET30B / Gene (production host): PTB DOMAIN OF IRS-1 / Production host: Skeleton / Plasmid: PET30B / Gene (production host): PTB DOMAIN OF IRS-1 / Production host:   Escherichia coli (E. coli) / References: UniProt: P35568 Escherichia coli (E. coli) / References: UniProt: P35568 |
|---|---|
| #2: Protein/peptide | Mass: 1241.311 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Production host: Homo sapiens (human) / Production host:   Escherichia coli (E. coli) / References: UniProt: P24394 Escherichia coli (E. coli) / References: UniProt: P24394 |
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR |
|---|
- Sample preparation
Sample preparation
Crystal grow | *PLUS Method: other / Details: NMR |
|---|
- Processing
Processing
| Software |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| NMR software | Name:  X-PLOR / Developer: BRUNGER / Classification: refinement X-PLOR / Developer: BRUNGER / Classification: refinement | ||||||||
| Refinement | Software ordinal: 1 Details: SET OF IDEAL BOND LENGTHS AND ANGLES USED DURING REFINEMENT: PARALLHDG.PRO IN X-PLOR. | ||||||||
| NMR ensemble | Conformers submitted total number: 1 |
 Movie
Movie Controller
Controller











 PDBj
PDBj












