+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
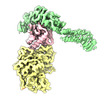
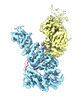
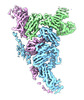
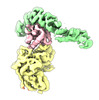
| タイトル | Structure of the yeast TRAPPII-Rab11/Ypt32 complex in the closed/closed state (composite structure) | |||||||||
 マップデータ マップデータ | Composite map of TRAPPII-Ypt32 symmetric closed/closed dimer | |||||||||
 試料 試料 |
| |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報Anchoring of the basal body to the plasma membrane / beta-glucan biosynthetic process / RAB geranylgeranylation / TRAPPI protein complex / RAB GEFs exchange GTP for GDP on RABs / TRAPPII protein complex / TRAPPIII protein complex /  TRAPP complex / early endosome to Golgi transport / COPII-mediated vesicle transport ...Anchoring of the basal body to the plasma membrane / beta-glucan biosynthetic process / RAB geranylgeranylation / TRAPPI protein complex / RAB GEFs exchange GTP for GDP on RABs / TRAPPII protein complex / TRAPPIII protein complex / TRAPP complex / early endosome to Golgi transport / COPII-mediated vesicle transport ...Anchoring of the basal body to the plasma membrane / beta-glucan biosynthetic process / RAB geranylgeranylation / TRAPPI protein complex / RAB GEFs exchange GTP for GDP on RABs / TRAPPII protein complex / TRAPPIII protein complex /  TRAPP complex / early endosome to Golgi transport / COPII-mediated vesicle transport / cytoplasm to vacuole targeting by the Cvt pathway / intra-Golgi vesicle-mediated transport / protein localization to phagophore assembly site / cellular bud neck / cis-Golgi network / protein-containing complex localization / phagophore assembly site / TRAPP complex / early endosome to Golgi transport / COPII-mediated vesicle transport / cytoplasm to vacuole targeting by the Cvt pathway / intra-Golgi vesicle-mediated transport / protein localization to phagophore assembly site / cellular bud neck / cis-Golgi network / protein-containing complex localization / phagophore assembly site /  retrograde transport, endosome to Golgi / cis-Golgi network membrane / retrograde transport, endosome to Golgi / cis-Golgi network membrane /  エキソサイトーシス / positive regulation of macroautophagy / chromosome organization / endoplasmic reticulum to Golgi vesicle-mediated transport / vesicle-mediated transport / Neutrophil degranulation / cell wall organization / エキソサイトーシス / positive regulation of macroautophagy / chromosome organization / endoplasmic reticulum to Golgi vesicle-mediated transport / vesicle-mediated transport / Neutrophil degranulation / cell wall organization /  オートファジー / オートファジー /  ゴルジ体 / recycling endosome / ゴルジ体 / recycling endosome /  オートファジー / オートファジー /  protein transport / protein-containing complex assembly / mitochondrial outer membrane / protein transport / protein-containing complex assembly / mitochondrial outer membrane /  エンドソーム / エンドソーム /  エンドソーム / エンドソーム /  ゴルジ体 / ゴルジ体 /  GTPase activity / GTP binding / GTPase activity / GTP binding /  ゴルジ体 / ゴルジ体 /  小胞体 / 小胞体 /  細胞核 / 細胞核 /  細胞質基質 / 細胞質基質 /  細胞質 細胞質類似検索 - 分子機能 | |||||||||
| 生物種 |   Saccharomyces cerevisiae (パン酵母) / Saccharomyces cerevisiae (パン酵母) /   Baker's yeast (パン酵母) / Baker's yeast (パン酵母) /   baker's yeast (パン酵母) baker's yeast (パン酵母) | |||||||||
| 手法 |  単粒子再構成法 / 単粒子再構成法 /  クライオ電子顕微鏡法 / 解像度: 3.7 Å クライオ電子顕微鏡法 / 解像度: 3.7 Å | |||||||||
 データ登録者 データ登録者 | Bagde SR / Fromme JC | |||||||||
| 資金援助 |  米国, 2件 米国, 2件
| |||||||||
 引用 引用 |  ジャーナル: Sci Adv / 年: 2022 ジャーナル: Sci Adv / 年: 2022タイトル: Structure of a TRAPPII-Rab11 activation intermediate reveals GTPase substrate selection mechanisms. 著者: Saket R Bagde / J Christopher Fromme /  要旨: Rab1 and Rab11 are essential regulators of the eukaryotic secretory and endocytic recycling pathways. The transport protein particle (TRAPP) complexes activate these guanosine triphosphatases via ...Rab1 and Rab11 are essential regulators of the eukaryotic secretory and endocytic recycling pathways. The transport protein particle (TRAPP) complexes activate these guanosine triphosphatases via nucleotide exchange using a shared set of core subunits. The basal specificity of the TRAPP core is toward Rab1, yet the TRAPPII complex is specific for Rab11. A steric gating mechanism has been proposed to explain TRAPPII counterselection against Rab1. Here, we present cryo-electron microscopy structures of the 22-subunit TRAPPII complex from budding yeast, including a TRAPPII-Rab11 nucleotide exchange intermediate. The Trs130 subunit provides a "leg" that positions the active site distal to the membrane surface, and this leg is required for steric gating. The related TRAPPIII complex is unable to activate Rab11 because of a repulsive interaction, which TRAPPII surmounts using the Trs120 subunit as a "lid" to enclose the active site. TRAPPII also adopts an open conformation enabling Rab11 to access and exit from the active site chamber. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_26254.map.gz emd_26254.map.gz | 28 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-26254-v30.xml emd-26254-v30.xml emd-26254.xml emd-26254.xml | 34.9 KB 34.9 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_26254.png emd_26254.png | 84.3 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26254 http://ftp.pdbj.org/pub/emdb/structures/EMD-26254 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26254 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26254 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  7u05MC  7u06C C: 同じ文献を引用 ( M: このマップから作成された原子モデル |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_26254.map.gz / 形式: CCP4 / 大きさ: 244.1 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_26254.map.gz / 形式: CCP4 / 大きさ: 244.1 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Composite map of TRAPPII-Ypt32 symmetric closed/closed dimer | ||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.432 Å | ||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
- 試料の構成要素
試料の構成要素
+全体 : TRAPPII complex bound to Rab11/Ypt32
+超分子 #1: TRAPPII complex bound to Rab11/Ypt32
+分子 #1: Trafficking protein particle complex II-specific subunit 130
+分子 #2: Trafficking protein particle complex II-specific subunit 65
+分子 #3: TRAPP-associated protein TCA17
+分子 #4: Trafficking protein particle complex subunit 33
+分子 #5: Trafficking protein particle complex subunit BET3
+分子 #6: Trafficking protein particle complex subunit BET5
+分子 #7: Trafficking protein particle complex subunit 23
+分子 #8: Trafficking protein particle complex subunit 31
+分子 #9: Trafficking protein particle complex subunit 20
+分子 #10: GTP-binding protein YPT32/YPT11
+分子 #11: Trafficking protein particle complex II-specific subunit 130
+分子 #12: Trafficking protein particle complex II-specific subunit 120
+分子 #13: Trafficking protein particle complex II-specific subunit 65
+分子 #14: PALMITIC ACID
-実験情報
-構造解析
| 手法 |  クライオ電子顕微鏡法 クライオ電子顕微鏡法 |
|---|---|
 解析 解析 |  単粒子再構成法 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 5.6 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 緩衝液 | pH: 8 構成要素:
| ||||||||||||
| グリッド | モデル: UltrAuFoil R1.2/1.3 / 材質: GOLD / メッシュ: 300 / 支持フィルム - 材質: GOLD / 支持フィルム - トポロジー: HOLEY / 支持フィルム - Film thickness: 50.0 nm / 前処理 - タイプ: GLOW DISCHARGE / 前処理 - 雰囲気: AIR | ||||||||||||
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % / チャンバー内温度: 277 K / 装置: FEI VITROBOT MARK IV 詳細: The sample was incubated on the grid for 10 seconds followed by blotting for 5 seconds before plunging in liquid ethane.. |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TALOS ARCTICA |
|---|---|
| 電子線 | 加速電圧: 200 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | C2レンズ絞り径: 50.0 µm / 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / 最大 デフォーカス(公称値): 2.5 µm / 最小 デフォーカス(公称値): 0.8 µm / 倍率(公称値): 63000 Bright-field microscopy / Cs: 2.7 mm / 最大 デフォーカス(公称値): 2.5 µm / 最小 デフォーカス(公称値): 0.8 µm / 倍率(公称値): 63000 |
| 特殊光学系 | エネルギーフィルター - 名称: GIF Bioquantum / エネルギーフィルター - スリット幅: 20 eV |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER ホルダー冷却材: NITROGEN |
| 撮影 | フィルム・検出器のモデル: GATAN K3 BIOQUANTUM (6k x 4k) デジタル化 - サイズ - 横: 5760 pixel / デジタル化 - サイズ - 縦: 4092 pixel / 撮影したグリッド数: 3 / 実像数: 4998 / 平均露光時間: 3.5 sec. / 平均電子線量: 53.0 e/Å2 / 詳細: Images were collected as 50 frame movies. |
| 実験機器 |  モデル: Talos Arctica / 画像提供: FEI Company |
- 画像解析
画像解析
| 粒子像選択 | 選択した数: 979187 | ||||||
|---|---|---|---|---|---|---|---|
| CTF補正 | ソフトウェア:
| ||||||
| 初期モデル | モデルのタイプ: INSILICO MODEL In silico モデル: Used Ab-initio Reconstruction job in cryoSPARC to generate starting map. | ||||||
| 初期 角度割当 | タイプ: MAXIMUM LIKELIHOOD / ソフトウェア - 名称: cryoSPARC (ver. 3.2.0) | ||||||
| 最終 角度割当 | タイプ: MAXIMUM LIKELIHOOD / ソフトウェア - 名称: RELION (ver. 3.1) | ||||||
| 最終 再構成 | 想定した対称性 - 点群: C2 (2回回転対称 ) / 解像度のタイプ: BY AUTHOR / 解像度: 3.7 Å / 解像度の算出法: FSC 0.143 CUT-OFF / ソフトウェア - 名称: PHENIX ) / 解像度のタイプ: BY AUTHOR / 解像度: 3.7 Å / 解像度の算出法: FSC 0.143 CUT-OFF / ソフトウェア - 名称: PHENIX詳細: The composite map was generated by combining the consensus dimer map (EMD-26221) with focused refinement maps deposited in the entries EMD-26223, EMD-26224. EMD-26225, EMD-26226, EMD-26227, ...詳細: The composite map was generated by combining the consensus dimer map (EMD-26221) with focused refinement maps deposited in the entries EMD-26223, EMD-26224. EMD-26225, EMD-26226, EMD-26227, EMD-26228, EMD-26229, EMD-26230, EMD-26231 and EMD-26232 using Combine Focused Maps in Phenix. 使用した粒子像数: 369488 |
 ムービー
ムービー コントローラー
コントローラー