[English] 日本語
 Yorodumi
Yorodumi- EMDB-26255: Structure of the yeast TRAPPII-Rab11/Ypt32 complex in the closed/... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the yeast TRAPPII-Rab11/Ypt32 complex in the closed/open state (composite structure) | |||||||||
 Map data Map data | composite map of TRAPPII-Ypt32 dimer in the closed/open state | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | complex / GTPase / Guanosine Exchange Factor / GEF / PROTEIN TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationAnchoring of the basal body to the plasma membrane / beta-glucan biosynthetic process / TRAPPI protein complex / RAB geranylgeranylation / RAB GEFs exchange GTP for GDP on RABs / TRAPPII protein complex / TRAPPIII protein complex / TRAPP complex / early endosome to Golgi transport / cytoplasm to vacuole targeting by the Cvt pathway ...Anchoring of the basal body to the plasma membrane / beta-glucan biosynthetic process / TRAPPI protein complex / RAB geranylgeranylation / RAB GEFs exchange GTP for GDP on RABs / TRAPPII protein complex / TRAPPIII protein complex / TRAPP complex / early endosome to Golgi transport / cytoplasm to vacuole targeting by the Cvt pathway / cis-Golgi network membrane / COPII-mediated vesicle transport / protein localization to phagophore assembly site / cellular bud neck / cis-Golgi network / intra-Golgi vesicle-mediated transport / phagophore assembly site / retrograde transport, endosome to Golgi / cellular response to nitrogen starvation / protein-containing complex localization / exocytosis / positive regulation of macroautophagy / chromosome organization / endoplasmic reticulum to Golgi vesicle-mediated transport / vesicle-mediated transport / Neutrophil degranulation / macroautophagy / trans-Golgi network / cell wall organization / recycling endosome / autophagy / protein transport / protein-containing complex assembly / early endosome / mitochondrial outer membrane / endosome / Golgi membrane / GTPase activity / GTP binding / endoplasmic reticulum / Golgi apparatus / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Bagde SR / Fromme JC | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Structure of a TRAPPII-Rab11 activation intermediate reveals GTPase substrate selection mechanisms. Authors: Saket R Bagde / J Christopher Fromme /  Abstract: Rab1 and Rab11 are essential regulators of the eukaryotic secretory and endocytic recycling pathways. The transport protein particle (TRAPP) complexes activate these guanosine triphosphatases via ...Rab1 and Rab11 are essential regulators of the eukaryotic secretory and endocytic recycling pathways. The transport protein particle (TRAPP) complexes activate these guanosine triphosphatases via nucleotide exchange using a shared set of core subunits. The basal specificity of the TRAPP core is toward Rab1, yet the TRAPPII complex is specific for Rab11. A steric gating mechanism has been proposed to explain TRAPPII counterselection against Rab1. Here, we present cryo-electron microscopy structures of the 22-subunit TRAPPII complex from budding yeast, including a TRAPPII-Rab11 nucleotide exchange intermediate. The Trs130 subunit provides a "leg" that positions the active site distal to the membrane surface, and this leg is required for steric gating. The related TRAPPIII complex is unable to activate Rab11 because of a repulsive interaction, which TRAPPII surmounts using the Trs120 subunit as a "lid" to enclose the active site. TRAPPII also adopts an open conformation enabling Rab11 to access and exit from the active site chamber. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26255.map.gz emd_26255.map.gz | 25.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26255-v30.xml emd-26255-v30.xml emd-26255.xml emd-26255.xml | 38.8 KB 38.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26255.png emd_26255.png | 192 KB | ||
| Filedesc metadata |  emd-26255.cif.gz emd-26255.cif.gz | 10.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26255 http://ftp.pdbj.org/pub/emdb/structures/EMD-26255 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26255 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26255 | HTTPS FTP |
-Related structure data
| Related structure data |  7u06MC  7u05C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26255.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26255.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | composite map of TRAPPII-Ypt32 dimer in the closed/open state | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.432 Å | ||||||||||||||||||||||||||||||||||||
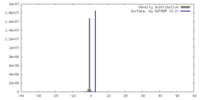
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
+Entire : TRAPPII complex bound to Rab11/Ypt32
+Supramolecule #1: TRAPPII complex bound to Rab11/Ypt32
+Macromolecule #1: Trafficking protein particle complex II-specific subunit 65
+Macromolecule #2: TRAPP-associated protein TCA17
+Macromolecule #3: Trafficking protein particle complex subunit 33
+Macromolecule #4: Trafficking protein particle complex subunit BET3
+Macromolecule #5: Trafficking protein particle complex subunit BET5
+Macromolecule #6: Trafficking protein particle complex subunit 23
+Macromolecule #7: Trafficking protein particle complex subunit 31
+Macromolecule #8: Trafficking protein particle complex subunit 20
+Macromolecule #9: GTP-binding protein YPT32/YPT11
+Macromolecule #10: Trafficking protein particle complex II-specific subunit 130
+Macromolecule #11: Trafficking protein particle complex II-specific subunit 120
+Macromolecule #12: Trafficking protein particle complex II-specific subunit 130
+Macromolecule #13: Trafficking protein particle complex II-specific subunit 65
+Macromolecule #14: PALMITIC ACID
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5.6 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Support film - Film thickness: 50 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: The sample was incubated on the grid for 10 seconds followed by blotting for 5 seconds before plunging in liquid ethane.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 3 / Number real images: 4998 / Average exposure time: 3.5 sec. / Average electron dose: 53.0 e/Å2 / Details: Images were collected as 50 frame movies. |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 63000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Details | Model was rebuilt in coot and refined using phenix.real_space_refine. | ||||||||
| Refinement | Space: REAL / Protocol: OTHER | ||||||||
| Output model |  PDB-7u06: |
 Movie
Movie Controller
Controller





















































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)