+ Open data
Open data
- Basic information
Basic information
| Entry | Database: SASBDB / ID: SASDBC4 |
|---|---|
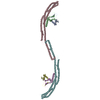
 Sample Sample | Plakin domain of Human plectin (spectrin repeats: SR3-SR9)
|
| Function / homology | Isoform 2 of Plectin Function and homology information Function and homology information |
| Biological species |  Homo sapiens (human) Homo sapiens (human) |
 Citation Citation |  Journal: J Biol Chem / Year: 2016 Journal: J Biol Chem / Year: 2016Title: The Structure of the Plakin Domain of Plectin Reveals an Extended Rod-like Shape. Authors: Esther Ortega / José A Manso / Rubén M Buey / Ana M Carballido / Arturo Carabias / Arnoud Sonnenberg / José M de Pereda /   Abstract: Plakins are large multi-domain proteins that interconnect cytoskeletal structures. Plectin is a prototypical plakin that tethers intermediate filaments to membrane-associated complexes. Most plakins ...Plakins are large multi-domain proteins that interconnect cytoskeletal structures. Plectin is a prototypical plakin that tethers intermediate filaments to membrane-associated complexes. Most plakins contain a plakin domain formed by up to nine spectrin repeats (SR1-SR9) and an SH3 domain. The plakin domains of plectin and other plakins harbor binding sites for junctional proteins. We have combined x-ray crystallography with small angle x-ray scattering (SAXS) to elucidate the structure of the plakin domain of plectin, extending our previous analysis of the SR1 to SR5 region. Two crystal structures of the SR5-SR6 region allowed us to characterize its uniquely wide inter-repeat conformational variability. We also report the crystal structures of the SR7-SR8 region, refined to 1.8 Å, and the SR7-SR9 at lower resolution. The SR7-SR9 region, which is conserved in all other plakin domains, forms a rigid segment stabilized by uniquely extensive inter-repeat contacts mediated by unusually long helices in SR8 and SR9. Using SAXS we show that in solution the SR3-SR6 and SR7-SR9 regions are rod-like segments and that SR3-SR9 of plectin has an extended shape with a small central kink. Other plakins, such as bullous pemphigoid antigen 1 and microtubule and actin cross-linking factor 1, are likely to have similar extended plakin domains. In contrast, desmoplakin has a two-segment structure with a central flexible hinge. The continuous versus segmented structures of the plakin domains of plectin and desmoplakin give insight into how different plakins might respond to tension and transmit mechanical signals. |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
-Models
| Model #496 |  Type: dummy / Software: DAMMIF / Radius of dummy atoms: 6.50 A / Chi-square value: 1.509 / P-value: 0.000090  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
|---|
- Sample
Sample
 Sample Sample | Name: Plakin domain of Human plectin (spectrin repeats: SR3-SR9) Specimen concentration: 0.92-14.70 |
|---|---|
| Buffer | Name: sodium phosphate / Concentration: 20.00 mM / pH: 7.5 / Composition: 150 mM NaCl, 5% glycerol, 3 mM DTT |
| Entity #321 | Name: Plectin / Type: protein Description: Plakin domain fragment of Human plectin encompassing spectrin repeats SR3-SR9 Formula weight: 95.573 / Num. of mol.: 1 / Source: Homo sapiens / References: UniProt: Q15149-2 Sequence: GSHMELEDST LRYLQDLLAW VEENQHRVDG AEWGVDLPSV EAQLGSHRGL HQSIEEFRAK IERARSDEGQ LSPATRGAYR DCLGRLDLQY AKLLNSSKAR LRSLESLHSF VAAATKELMW LNEKEEEEVG FDWSDRNTNM TAKKESYSAL MRELELKEKK IKELQNAGDR ...Sequence: GSHMELEDST LRYLQDLLAW VEENQHRVDG AEWGVDLPSV EAQLGSHRGL HQSIEEFRAK IERARSDEGQ LSPATRGAYR DCLGRLDLQY AKLLNSSKAR LRSLESLHSF VAAATKELMW LNEKEEEEVG FDWSDRNTNM TAKKESYSAL MRELELKEKK IKELQNAGDR LLREDHPARP TVESFQAALQ TQWSWMLQLC CCIEAHLKEN AAYFQFFSDV REAEGQLQKL QEALRRKYSC DRSATVTRLE DLLQDAQDEK EQLNEYKGHL SGLAKRAKAV VQLKPRHPAH PMRGRLPLLA VCDYKQVEVT VHKGDECQLV GPAQPSHWKV LSSSGSEAAV PSVCFLVPPP NQEAQEAVTR LEAQHQALVT LWHQLHVDMK SLLAWQSLRR DVQLIRSWSL ATFRTLKPEE QRQALHSLEL HYQAFLRDSQ DAGGFGPEDR LMAEREYGSC SHHYQQLLQS LEQGAQEESR CQRCISELKD IRLQLEACET RTVHRLRLPL DKEPARECAQ RIAEQQKAQA EVEGLGKGVA RLSAEAEKVL ALPEPSPAAP TLRSELELTL GKLEQVRSLS AIYLEKLKTI SLVIRGTQGA EEVLRAHEEQ LKEAQAVPAT LPELEATKAS LKKLRAQAEA QQPTFDALRD ELRGAQEVGE RLQQRHGERD VEVERWRERV AQLLERWQAV LAQTDVRQRE LEQLGRQLRY YRESADPLGA WLQDARRRQE QIQAMPLADS QAVREQLRQE QALLEEIERH GEKVEECQRF AKQYINAIKD YELQLVTYKA QLEPVASPAK KPKVQSGSES VIQEYVDLRT HYSELTTLTS QYIKFISETL RRME |
-Experimental information
| Beam | Instrument name: PETRA III P12 / City: Hamburg / 国: Germany  / Type of source: X-ray synchrotron / Wavelength: 0.12 Å / Dist. spec. to detc.: 3.1 mm / Type of source: X-ray synchrotron / Wavelength: 0.12 Å / Dist. spec. to detc.: 3.1 mm | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 2M | ||||||||||||||||||
| Scan |
| ||||||||||||||||||
| Distance distribution function P(R) |
| ||||||||||||||||||
| Result |
|
 Movie
Movie Controller
Controller


 SASDBC4
SASDBC4