+ Open data
Open data
- Basic information
Basic information
| Entry | Database: SASBDB / ID: SASDE56 |
|---|---|
 Sample Sample | Human ATP-citrate synthase (ACLY) in HBS + Citrate + Coenzyme-A
|
| Function / homology | ATP citrate synthase / Isoform 1 of ATP-citrate synthase Function and homology information Function and homology information |
| Biological species |  Homo sapiens (human) Homo sapiens (human) |
 Citation Citation |  Date: 2019 Apr 3 Date: 2019 Apr 3Title: Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle Authors: Verschueren K / Blanchet C / Felix J / Dansercoer A / De Vos D / Bloch Y / Van Beeumen J / Svergun D / Gutsche I / Savvides S |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
-Models
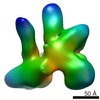
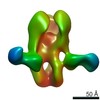
| Model #2506 |  Type: atomic / Symmetry: P222 / Chi-square value: 7.25  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
|---|---|
| Model #2875 |  Type: atomic / Radius of dummy atoms: 1.90 A / Chi-square value: 2.75656623518009  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
- Sample
Sample
 Sample Sample | Name: Human ATP-citrate synthase (ACLY) in HBS + Citrate + Coenzyme-A Specimen concentration: 24 mg/ml |
|---|---|
| Buffer | Name: 20mM HEPES, 150mM NaCl, 50mM Tris, 20mM citrate, 2mM CoA pH: 7.2 |
| Entity #1335 | Type: protein / Description: ATP-citrate synthase / Formula weight: 114.58 / Num. of mol.: 4 / Source: Homo sapiens / References: UniProt: P53396-1 Sequence: MSAKAISEQT GKELLYKFIC TTSAIQNRFK YARVTPDTDW ARLLQDHPWL LSQNLVVKPD QLIKRRGKLG LVGVNLTLDG VKSWLKPRLG QEATVGKATG FLKNFLIEPF VPHSQAEEFY VCIYATREGD YVLFHHEGGV DVGDVDAKAQ KLLVGVDEKL NPEDIKKHLL ...Sequence: MSAKAISEQT GKELLYKFIC TTSAIQNRFK YARVTPDTDW ARLLQDHPWL LSQNLVVKPD QLIKRRGKLG LVGVNLTLDG VKSWLKPRLG QEATVGKATG FLKNFLIEPF VPHSQAEEFY VCIYATREGD YVLFHHEGGV DVGDVDAKAQ KLLVGVDEKL NPEDIKKHLL VHAPEDKKEI LASFISGLFN FYEDLYFTYL EINPLVVTKD GVYVLDLAAK VDATADYICK VKWGDIEFPP PFGREAYPEE AYIADLDAKS GASLKLTLLN PKGRIWTMVA GGGASVVYSD TICDLGGVNE LANYGEYSGA PSEQQTYDYA KTILSLMTRE KHPDGKILII GGSIANFTNV AATFKGIVRA IRDYQGPLKE HEVTIFVRRG GPNYQEGLRV MGEVGKTTGI PIHVFGTETH MTAIVGMALG HRPIPGKSTT LFSRHTKAIV WGMQTRAVQG MLDFDYVCSR DEPSVAAMVY PFTGDHKQKF YWGHKEILIP VFKNMADAMR KHPEVDVLIN FASLRSAYDS TMETMNYAQI RTIAIIAEGI PEALTRKLIK KADQKGVTII GPATVGGIKP GCFKIGNTGG MLDNILASKL YRPGSVAYVS RSGGMSNELN NIISRTTDGV YEGVAIGGDR YPGSTFMDHV LRYQDTPGVK MIVVLGEIGG TEEYKICRGI KEGRLTKPIV CWCIGTCATM FSSEVQFGHA GACANQASET AVAKNQALKE AGVFVPRSFD ELGEIIQSVY EDLVANGVIV PAQEVPPPTV PMDYSWAREL GLIRKPASFM TSICDERGQE LIYAGMPITE VFKEEMGIGG VLGLLWFQKR LPKYSCQFIE MCLMVTADHG PAVSGAHNTI ICARAGKDLV SSLTSGLLTI GDRFGGALDA AAKMFSKAFD SGIIPMEFVN KMKKEGKLIM GIGHRVKSIN NPDMRVQILK DYVRQHFPAT PLLDYALEVE KITTSKKPNL ILNVDGLIGV AFVDMLRNCG SFTREEADEY IDIGALNGIF VLGRSMGFIG HYLDQKRLKQ GLYRHPWDDI SYVLPEHMSM |
-Experimental information
| Beam | Instrument name: PETRA III EMBL P12 / City: Hamburg / 国: Germany  / Type of source: X-ray synchrotron / Wavelength: 0.124 Å / Dist. spec. to detc.: 3.1 mm / Type of source: X-ray synchrotron / Wavelength: 0.124 Å / Dist. spec. to detc.: 3.1 mm | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 6M | ||||||||||||||||||||||||||||||
| Scan | Measurement date: May 5, 2018 / Storage temperature: 20 °C / Cell temperature: 20 °C / Exposure time: 1 sec. / Number of frames: 900 / Unit: 1/nm /
| ||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| ||||||||||||||||||||||||||||||
| Result |
|
 Movie
Movie Controller
Controller


 SASDE56
SASDE56