[English] 日本語
 Yorodumi
Yorodumi- SASDAE7: Structure of a complex between full length and truncated CS74L en... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: SASBDB / ID: SASDAE7 |
|---|---|
 Sample Sample | Structure of a complex between full length and truncated CS74L endolysin
|
| Function / homology | : / Ami_3 / N-acetylmuramoyl-L-alanine amidase, catalytic domain / N-acetylmuramoyl-L-alanine amidase / N-acetylmuramoyl-L-alanine amidase activity / viral release from host cell by cytolysis / peptidoglycan catabolic process / Endolysin CS74L Function and homology information Function and homology information |
| Biological species |  Clostridium phage phi8074-B1 (virus) Clostridium phage phi8074-B1 (virus) |
 Citation Citation |  Journal: J Biol Chem / Year: 2016 Journal: J Biol Chem / Year: 2016Title: Crystal Structure of the CTP1L Endolysin Reveals How Its Activity Is Regulated by a Secondary Translation Product. Authors: Matthew Dunne / Stefan Leicht / Boris Krichel / Haydyn D T Mertens / Andrew Thompson / Jeroen Krijgsveld / Dmitri I Svergun / Natalia Gómez-Torres / Sonia Garde / Charlotte Uetrecht / Arjan ...Authors: Matthew Dunne / Stefan Leicht / Boris Krichel / Haydyn D T Mertens / Andrew Thompson / Jeroen Krijgsveld / Dmitri I Svergun / Natalia Gómez-Torres / Sonia Garde / Charlotte Uetrecht / Arjan Narbad / Melinda J Mayer / Rob Meijers /     Abstract: Bacteriophages produce endolysins, which lyse the bacterial host cell to release newly produced virions. The timing of lysis is regulated and is thought to involve the activation of a molecular ...Bacteriophages produce endolysins, which lyse the bacterial host cell to release newly produced virions. The timing of lysis is regulated and is thought to involve the activation of a molecular switch. We present a crystal structure of the activated endolysin CTP1L that targets Clostridium tyrobutyricum, consisting of a complex between the full-length protein and an N-terminally truncated C-terminal cell wall binding domain (CBD). The truncated CBD is produced through an internal translation start site within the endolysin gene. Mutants affecting the internal translation site change the oligomeric state of the endolysin and reduce lytic activity. The activity can be modulated by reconstitution of the full-length endolysin-CBD complex with free CBD. The same oligomerization mechanism applies to the CD27L endolysin that targets Clostridium difficile and the CS74L endolysin that targets Clostridium sporogenes. When the CTP1L endolysin gene is introduced into the commensal bacterium Lactococcus lactis, the truncated CBD is also produced, showing that the alternative start codon can be used in other bacterial species. The identification of a translational switch affecting oligomerization presented here has implications for the design of effective endolysins for the treatment of bacterial infections. |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
-Models
| Model #257 |  Type: dummy / Radius of dummy atoms: 3.00 A / Chi-square value: 0.775  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
|---|---|
| Model #258 |  Type: mix / Software: (2.0) / Radius of dummy atoms: 1.90 A / Comment: Ensemble Component: Heterotetramer (compact) / Chi-square value: 0.664 / P-value: 0.051000  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
| Model #259 |  Type: mix / Radius of dummy atoms: 1.90 A / Comment: Ensemble Component: Heterotetramer (extended) / Chi-square value: 0.664 / P-value: 0.051000  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
| Model #260 | 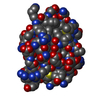 Type: mix / Software: (2.0) / Radius of dummy atoms: 1.90 A / Comment: Ensemble Component: Free CTD of endolysin / Chi-square value: 0.664 / P-value: 0.051000  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
- Sample
Sample
 Sample Sample | Name: Structure of a complex between full length and truncated CS74L endolysin Specimen concentration: 1.00-6.80 |
|---|---|
| Buffer | Name: 20 mM HEPES / pH: 7.4 |
| Entity #156 | Name: CS74L / Type: protein / Description: Endolysin CS74L / Formula weight: 31.1 / Source: Clostridium phage phi8074-B1 / References: UniProt: I1TJX3 Sequence: MGSSHHHHHH SSGLVPRGSH MKIGIDMGHT LSGADYGVVG LRPESVLTRE VGTKVIYKLQ KLGHVVVNCT VDKASSVSES LYTRYYRANQ ANVDLFISIH FNATPGGTGT EVYTYAGRQL GEATRIRQEF KSLGLRDRGT KDGSGLAVIR NTKAKAMLVE CCFCDNPNDM ...Sequence: MGSSHHHHHH SSGLVPRGSH MKIGIDMGHT LSGADYGVVG LRPESVLTRE VGTKVIYKLQ KLGHVVVNCT VDKASSVSES LYTRYYRANQ ANVDLFISIH FNATPGGTGT EVYTYAGRQL GEATRIRQEF KSLGLRDRGT KDGSGLAVIR NTKAKAMLVE CCFCDNPNDM KLYNSESFSN AIVKGITGKL PNGESGNNNQ GGNKVKAVVI YNEGADRRGA EYLADYLNCP TISNSRTFDY SCVEHVYAVG GKKEQYTKYL KTLLSGANRY DTMQQILNFI NGGK |
-Experimental information
| Beam | Instrument name:  DORIS III X33 DORIS III X33  / City: Hamburg / 国: Germany / City: Hamburg / 国: Germany  / Shape: 0.6 / Type of source: X-ray synchrotron / Wavelength: 0.15 Å / Dist. spec. to detc.: 2.7 mm / Shape: 0.6 / Type of source: X-ray synchrotron / Wavelength: 0.15 Å / Dist. spec. to detc.: 2.7 mm | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 1M-W / Pixsize x: 0.172 mm | |||||||||||||||||||||
| Scan |
| |||||||||||||||||||||
| Distance distribution function P(R) |
| |||||||||||||||||||||
| Result |
|
 Movie
Movie Controller
Controller

 SASDAE7
SASDAE7