+ Open data
Open data
- Basic information
Basic information
| Entry | Database: SASBDB / ID: SASDCM6 |
|---|---|
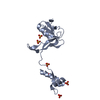
 Sample Sample | Small GTPase Rab5 conjugated with ubiquitin at K116
|
| Biological species |  Homo sapiens (human) Homo sapiens (human) |
 Citation Citation |  Journal: Elife / Year: 2017 Journal: Elife / Year: 2017Title: Site-specific monoubiquitination downregulates Rab5 by disrupting effector binding and guanine nucleotide conversion. Authors: Donghyuk Shin / Wooju Na / Ji-Hyung Lee / Gyuhee Kim / Jiseok Baek / Seok Hee Park / Cheol Yong Choi / Sangho Lee /  Abstract: Rab GTPases, which are involved in intracellular trafficking pathways, have recently been reported to be ubiquitinated. However, the functions of ubiquitinated Rab proteins remain unexplored. Here we ...Rab GTPases, which are involved in intracellular trafficking pathways, have recently been reported to be ubiquitinated. However, the functions of ubiquitinated Rab proteins remain unexplored. Here we show that Rab5 is monoubiquitinated on K116, K140, and K165. Upon co-transfection with ubiquitin, Rab5 exhibited abnormalities in endosomal localization and EGF-induced EGF receptor degradation. Rab5 K140R and K165R mutants restored these abnormalities, whereas K116R did not. We derived structural models of individual monoubiquitinated Rab5 proteins (mUbRab5s) by solution scattering and observed different conformational flexibilities in a site-specific manner. Structural analysis combined with biochemical data revealed that interactions with downstream effectors were impeded in mUbRab5, whereas GDP release and GTP loading activities were altered in mUbRab5. By contrast, mUbRab5 apparently had no effect. We propose a regulatory mechanism of Rab5 where monoubiquitination downregulates effector recruitment and GDP/GTP conversion in a site-specific manner. |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
-Models
| Model #1448 |  Type: mix / Radius of dummy atoms: 1.90 A / Chi-square value: 1.145  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
|---|---|
| Model #1449 |  Type: mix / Radius of dummy atoms: 1.90 A / Chi-square value: 1.145  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
| Model #1450 |  Type: dummy / Radius of dummy atoms: 1.80 A Comment: The lowest NSD model from damaver with 10 independent dammif runs Chi-square value: 1.316  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
- Sample
Sample
 Sample Sample | Name: Small GTPase Rab5 conjugated with ubiquitin at K116 / Specimen concentration: 0.40-2.90 |
|---|---|
| Buffer | Name: 50 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl2 / pH: 7.5 |
| Entity #760 | Name: mUbRab5K165 / Type: protein / Description: Monoubiquitinated Rab5 at K165 / Formula weight: 32.205 / Num. of mol.: 1 / Source: Homo sapiens Sequence: MQIFVKTLTG KTITLEVEPS DTIENVKAKI QDKEGIPPDQ QRLIFAGKQL EDGRTLSDYN IQKESTLHLV LRLRGGMASR GATRPNGPNT GNKICQFKLV LLGESAVGKS SLVLRFVKGQ FHEFQESTIG AAFLTQTVCL DDTTVKFEIW DTAGQERYHS LAPMYYRGAQ ...Sequence: MQIFVKTLTG KTITLEVEPS DTIENVKAKI QDKEGIPPDQ QRLIFAGKQL EDGRTLSDYN IQKESTLHLV LRLRGGMASR GATRPNGPNT GNKICQFKLV LLGESAVGKS SLVLRFVKGQ FHEFQESTIG AAFLTQTVCL DDTTVKFEIW DTAGQERYHS LAPMYYRGAQ AAIVVYDITN EESFARAKNW VKELQRQASP NIVIALSGNK ADLANKRAVD FQEAQSYADD NSLLFMETSA KTSMNVNEIF MAIAKKLPKN EPQNPGANSA RGRGVDLTEP TQPTRNQCCS N |
-Experimental information
| Beam | Instrument name: Pohang Accelerator Laboratory 4C / City: Pohang / 国: South Korea / Type of source: X-ray synchrotron / Wavelength: 0.124 Å / Dist. spec. to detc.: 3 mm | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: ADSC Quantum 315 / Type: CCD / Pixsize x: 315 mm | ||||||||||||
| Scan |
| ||||||||||||
| Distance distribution function P(R) |
| ||||||||||||
| Result |
|
 Movie
Movie Controller
Controller


 SASDCM6
SASDCM6