[English] 日本語
 Yorodumi
Yorodumi- PDB-8rkg: Crystal structure of tetrameric collagenase-cleaved Xenopus ZP2-N... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8rkg | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure of tetrameric collagenase-cleaved Xenopus ZP2-N2N3 (cleaved xZP2-N2N3) | ||||||||||||
 Components Components | (XlZPA protein) x 2 | ||||||||||||
 Keywords Keywords | CELL ADHESION / fertilization / egg-sperm interaction / gamete recognition / sperm receptor / extracellular matrix / egg coat / zona pellucida / glycoprotein / N-glycan / structural protein / ZP-N domain / block to polyspermy / post-fertilization cleavage / ovastacin / AlphaFold | ||||||||||||
| Function / homology |  Function and homology information Function and homology information | ||||||||||||
| Biological species | |||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.9 Å MOLECULAR REPLACEMENT / Resolution: 2.9 Å | ||||||||||||
 Authors Authors | Nishio, S. / de Sanctis, D. / Jovine, L. | ||||||||||||
| Funding support |  Sweden, 3items Sweden, 3items
| ||||||||||||
 Citation Citation |  Journal: Cell / Year: 2024 Journal: Cell / Year: 2024Title: ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat. Authors: Shunsuke Nishio / Chihiro Emori / Benjamin Wiseman / Dirk Fahrenkamp / Elisa Dioguardi / Sara Zamora-Caballero / Marcel Bokhove / Ling Han / Alena Stsiapanava / Blanca Algarra / Yonggang Lu ...Authors: Shunsuke Nishio / Chihiro Emori / Benjamin Wiseman / Dirk Fahrenkamp / Elisa Dioguardi / Sara Zamora-Caballero / Marcel Bokhove / Ling Han / Alena Stsiapanava / Blanca Algarra / Yonggang Lu / Mayo Kodani / Rachel E Bainbridge / Kayla M Komondor / Anne E Carlson / Michael Landreh / Daniele de Sanctis / Shigeki Yasumasu / Masahito Ikawa / Luca Jovine /     Abstract: Following the fertilization of an egg by a single sperm, the egg coat or zona pellucida (ZP) hardens and polyspermy is irreversibly blocked. These events are associated with the cleavage of the N- ...Following the fertilization of an egg by a single sperm, the egg coat or zona pellucida (ZP) hardens and polyspermy is irreversibly blocked. These events are associated with the cleavage of the N-terminal region (NTR) of glycoprotein ZP2, a major subunit of ZP filaments. ZP2 processing is thought to inactivate sperm binding to the ZP, but its molecular consequences and connection with ZP hardening are unknown. Biochemical and structural studies show that cleavage of ZP2 triggers its oligomerization. Moreover, the structure of a native vertebrate egg coat filament, combined with AlphaFold predictions of human ZP polymers, reveals that two protofilaments consisting of type I (ZP3) and type II (ZP1/ZP2/ZP4) components interlock into a left-handed double helix from which the NTRs of type II subunits protrude. Together, these data suggest that oligomerization of cleaved ZP2 NTRs extensively cross-links ZP filaments, rigidifying the egg coat and making it physically impenetrable to sperm. #1: Journal: Dev Biol / Year: 1986 Title: The vitelline envelope to fertilization envelope conversion in eggs of Xenopus laevis. Abstract: Fertilization of the Xenopus laevis egg causes the conversion of the vitelline envelope to the fertilization envelope, a change reflected in the loss of sperm penetrability of the egg and the ...Fertilization of the Xenopus laevis egg causes the conversion of the vitelline envelope to the fertilization envelope, a change reflected in the loss of sperm penetrability of the egg and the appearance of an electron-dense layer on the outer aspect of the fertilization envelope. As seen by one-dimensional gel electrophoresis, two components with molecular weights of 69,000 and 64,000 in the vitelline envelope were converted to 66,000 and 61,000 in the fertilization envelope. By two-dimensional gel electrophoresis, the components in the 69,000 and 64,000 molecular weight regions of the vitelline envelope were seen to shift to more basic isoelectric points upon conversion to the fertilization envelope. Peptide mapping by limited proteolysis suggested that the 69,000 and 64,000 molecular weight components shared the same polypeptide chains but the smaller glycoprotein lacked a carbohydrate side chain found on the larger species. Similar sites on each glycoprotein were affected when the vitelline envelope was converted to the fertilization envelope. No N-terminal amino acids could be identified on the envelope components, indicating that these glycoproteins have blocked N-termini. Ionophore A23187-activation of jellied eggs (but not dejellied eggs) caused the molecular weight changes in the absence of sperm. Thus, factors from the jelly and the cortical granules but not from sperm apparently are involved in the processing of the 69,000 and 64,000 molecular weight components. #2: Journal: J Cell Biol / Year: 1997 Title: Gamete interactions in Xenopus laevis: identification of sperm binding glycoproteins in the egg vitelline envelope. Authors: J Tian / H Gong / G H Thomsen / W J Lennarz /  Abstract: A quantitative assay was developed to study the interaction of Xenopus laevis sperm and eggs. Using this assay it was found that sperm bound in approximately equal numbers to the surface of both ...A quantitative assay was developed to study the interaction of Xenopus laevis sperm and eggs. Using this assay it was found that sperm bound in approximately equal numbers to the surface of both hemispheres of the unfertilized egg, but not to the surface of the fertilized egg. To understand the molecular basis of sperm binding to the egg vitelline envelope (VE), a competition assay was used and it was found that solubilized total VE proteins inhibited sperm-egg binding in a concentration-dependent manner. Individual VE proteins were then isolated and tested for their ability to inhibit sperm binding. Of the seven proteins in the VE, two related glycoproteins, gp69 and gp64, inhibited sperm-egg binding. Polyclonal antibody was prepared that specifically recognized gp69 and gp64. This gp69/64 specific antibody bound to the VE surface and blocked sperm binding, as well as fertilization. Moreover, agarose beads coated with gp69/64 showed high sperm binding activity, while beads coated with other VE proteins bound few sperm. Treatment of unfertilized eggs with crude collagenase resulted in proteolytic modification of only the gp69/64 components of the VE, and this modification abolished sperm-egg binding. Small glycopeptides generated by Pronase digestion of gp69/64 also inhibited sperm-egg binding and this inhibition was abolished by treatment of the glycopeptides with periodate. Based on these observations, we conclude that the gp69/64 glycoproteins in the egg vitelline envelope mediate sperm-egg binding, an initial step in Xenopus fertilization, and that the oligosaccharide chains of these glycoproteins may play a critical role in this process. #3: Journal: Dev Biol / Year: 1997 Title: Xenopus laevis sperm-egg adhesion is regulated by modifications in the sperm receptor and the egg vitelline envelope. Authors: J Tian / H Gong / G H Thomsen / W J Lennarz /  Abstract: The biochemical and ultrastructural changes in the envelope of the Xenopus laevis egg that occur during oviposition and fertilization have been thoroughly studied (Hedrick, J. L., and Nishihara, D. M. ...The biochemical and ultrastructural changes in the envelope of the Xenopus laevis egg that occur during oviposition and fertilization have been thoroughly studied (Hedrick, J. L., and Nishihara, D. M., Methods Cell Biol. 36, 231-247, 1991; Larabell, C. A., and Chandler, D. E., J. Electron Microsc. Tech. 17, 294-318, 1991). However, the biological significance of these changes with respect to gamete interaction has been unclear. In the current study, it was found that changes in the envelope are directly responsible for regulating sperm-egg adhesion, an initial step of fertilization. As a result of these transformations, sperm bind only to unfertilized oviposited eggs, not to oocytes or coelomic eggs. In addition, they do not bind to fertilized eggs. The molecular and cellular basis of the regulation of the sperm binding process was investigated in the context of our recent findings that two structurally related envelope glycoproteins, gp69/64, serve as sperm receptors during fertilization (Tian, J.-D., Gong, H., Thomsen, G. H., and Lennarz, W. J., J. Cell Biol. 136, 1099-1108, 1997). Although the purified gp69/64 glycoproteins isolated from the oocyte or coelomic egg envelopes exhibited sperm binding activity, when these proteins are part of the intact oocyte or coelomic egg envelopes, they are not accessible to either anti-gp69/64 antibodies or to sperm. During the conversion from the coelomic to the vitelline envelope, the gp69/64 sperm receptors become exposed on the surface, an event that correlates with proteolytic cleavage of gp43 and accompanying ultrastructural alterations in the envelope. Conversely, after fertilization, when the vitelline envelope of the egg is converted to the fertilization envelope of the zygote, limited proteolytic cleavage of the sperm receptor results in loss of sperm binding activity. In addition, formation of a fertilization layer on top of the structurally altered VE adds another physical block to sperm binding. These results provide new insights into structure-function relationships between envelope components of the anuran egg, and provide further evidence supporting the key role of gp69/64 as sperm receptors during X. laevis fertilization. #4: Journal: Proc Natl Acad Sci U S A / Year: 1999 Title: Xenopus laevis sperm receptor gp69/64 glycoprotein is a homolog of the mammalian sperm receptor ZP2. Authors: J Tian / H Gong / W J Lennarz /  Abstract: Little is known about sperm-binding proteins in the egg envelope of nonmammalian vertebrate species. We report here the molecular cloning and characterization of a recently identified sperm receptor ...Little is known about sperm-binding proteins in the egg envelope of nonmammalian vertebrate species. We report here the molecular cloning and characterization of a recently identified sperm receptor (gp69/64) in the Xenopus laevis egg vitelline envelope. Our data indicate that the gp69 and gp64 glycoproteins are two glycoforms of the receptor and have the same number of N-linked oligosaccharide chains but differ in the extent of O-glycosylation. The amino acid sequence of the receptor is closely related to that of the mouse zona pellucida protein ZP2. Most of the sequence conservation, including a ZP domain, a potential furin cleavage site, and a putative transmembrane domain are located in the C-terminal half of the receptor. Proteolytic cleavage of the gp69/64 protein by a cortical granule protease during fertilization removes 27 amino acid residues from the N terminus of gp69/64 and results in loss of sperm binding to the activated eggs. Similarly, we find that treatment of eggs with type I collagenase removes 31 residues from the N terminus of gp69/64 and has the same effect on sperm binding. The isolated and purified N terminus-truncated receptor protein is inactive as an inhibitor of sperm-egg binding. Earlier studies on the effect of Pronase digestion on receptor activity suggest that this N-terminal peptide may contain an O-linked glycan that is involved in the binding process. Based on these results and the findings on the primary structure of the receptor, a pathway for the maturation and secretion of gp69/64, as well as its inactivation following fertilization, is proposed. #5: Journal: Biochem Biophys Res Commun / Year: 2004 Title: Proteolysis of Xenopus laevis egg envelope ZPA triggers envelope hardening. Authors: Leann L Lindsay / Jerry L Hedrick /  Abstract: The egg envelope of most animal eggs is modified following fertilization, resulting in the prevention of polyspermy and hardening of the egg envelope. In frogs and mammals a prominent feature of ...The egg envelope of most animal eggs is modified following fertilization, resulting in the prevention of polyspermy and hardening of the egg envelope. In frogs and mammals a prominent feature of envelope modification is N-terminal proteolysis of the envelope glycoprotein ZPA. We have purified the ZPA protease from Xenopus laevis eggs and characterized it as a zinc metalloprotease. Proteolysis of isolated egg envelopes by the isolated protease resulted in envelope hardening. The N-terminal peptide fragment of ZPA remained disulfide bond linked to the ZPA glycoprotein moiety following proteolysis. We propose a mechanism for egg envelope hardening involving ZPA proteolysis by an egg metalloprotease as a triggering event followed by induction of global conformational changes in egg envelope glycoproteins. #6: Journal: Biol Cell / Year: 2008 Title: Cellular origin of the Bufo arenarum sperm receptor gp75, a ZP2 family member: its proteolysis after fertilization. Authors: Sonia L Scarpeci / Mercedes L Sanchez / Marcelo O Cabada /  Abstract: BACKGROUND INFORMATION: The egg envelope is an extracellular matrix that surrounds oocytes. In frogs and mammals, a prominent feature of envelope modification following fertilization is the N- ...BACKGROUND INFORMATION: The egg envelope is an extracellular matrix that surrounds oocytes. In frogs and mammals, a prominent feature of envelope modification following fertilization is the N-terminal proteolysis of the envelope glycoproteins, ZPA [ZP (zona pellucida) A]. It was proposed that ZPA N-terminal proteolysis leads to a conformational change in egg envelope glycoproteins, resulting in the prevention of polyspermy. Bufo arenarum VE (vitelline envelope) is made up of at least four glycoproteins: gp120 (glycoprotein 120), gp75, gp41 and gp38. The aim of the present study was to identify and characterize the baZPA (B. arenarum ZPA homologue). Also, our aim was to evaluate its integrity and functional significance during fertilization. RESULTS: VE components were labelled with FITC in order to study their sperm-binding capacity. The assay showed that gp75, gp41 and gp38 possess sperm-binding activity. We obtained a full-length cDNA ...RESULTS: VE components were labelled with FITC in order to study their sperm-binding capacity. The assay showed that gp75, gp41 and gp38 possess sperm-binding activity. We obtained a full-length cDNA of 2062 bp containing one ORF (open reading frame) with a sequence for 687 amino acids. The predicted amino acid sequence had close similarity to that of mammalian ZPA. This result indicates that gp75 is the baZPA. Antibodies raised against an N-terminal sequence recognized baZPA and inhibited sperm-baZPA extracted from VE binding. This protein does not induce the acrosome reaction in homologue sperm. Northern-blot studies indicated that the transcript is exclusively expressed in the ovary. In situ hybridization studies confirmed this and pointed to previtellogenic oocytes and follicle cells surrounding the oocyte as the source of the transcript. baZPA was cleaved during fertilization and the N-terminal peptide fragment remained disulfide bonded to the glycoprotein moiety following proteolysis. CONCLUSION: From the sequence analysis, it was possible to consider that gp75 is the baZPA. It is expressed by previtellogenic oocytes and follicle cells. Also, it can be considered as a sperm ...CONCLUSION: From the sequence analysis, it was possible to consider that gp75 is the baZPA. It is expressed by previtellogenic oocytes and follicle cells. Also, it can be considered as a sperm receptor that undergoes N-terminal proteolysis during fertilization. The N-terminal peptide could be necessary for sperm binding. #7: Journal: Int J Dev Biol / Year: 2008 Title: Anuran and pig egg zona pellucida glycoproteins in fertilization and early development. Authors: Jerry L Hedrick /  Abstract: The envelope surrounding the eggs of all animals has many biological functions in fertilization and development. This review focuses on the anuran egg envelope in terms of its biochemistry, ...The envelope surrounding the eggs of all animals has many biological functions in fertilization and development. This review focuses on the anuran egg envelope in terms of its biochemistry, biophysics, structural biology and function in sperm-egg interactions and early development (embryo hatching). Egg envelopes from Xenopus laevis are among the most studied of the anurans, and are the central theme of this review. Comparisons of Xenopus laevis envelopes with those of other anurans and with pig egg envelopes are also included. This article presents historical as well as contemporary comparative perspectives on molecular and cellular mechanisms of sperm-egg envelope binding, block to polyspermy, envelope hardening, and hatching. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8rkg.cif.gz 8rkg.cif.gz | 344.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8rkg.ent.gz pdb8rkg.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  8rkg.json.gz 8rkg.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/rk/8rkg https://data.pdbj.org/pub/pdb/validation_reports/rk/8rkg ftp://data.pdbj.org/pub/pdb/validation_reports/rk/8rkg ftp://data.pdbj.org/pub/pdb/validation_reports/rk/8rkg | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  8bquC  8rkeC 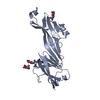 8rkfC  8rkhC  8rkiC C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||
| 2 | 
| ||||||||||||
| Unit cell |
|
- Components
Components
-Protein/peptide / Protein , 2 types, 16 molecules LMNOPQRSABCDEFGH
| #1: Protein/peptide | Mass: 3356.714 Da / Num. of mol.: 8 Source method: isolated from a genetically manipulated source Details: The protein consists of two polypeptide fragments, generated by collagenase cleavage between P160 and V161, that remain covalently linked via a disulfide bond between C139 and C244 Source: (gene. exp.)  Homo sapiens (human) / References: UniProt: A1L3D9 Homo sapiens (human) / References: UniProt: A1L3D9#2: Protein | Mass: 21035.756 Da / Num. of mol.: 8 Source method: isolated from a genetically manipulated source Details: The protein consists of two polypeptide fragments, generated by collagenase cleavage between P160 and V161, that remain covalently linked via a disulfide bond between C139 and C244 Source: (gene. exp.)  Homo sapiens (human) / References: UniProt: A1L3D9 Homo sapiens (human) / References: UniProt: A1L3D9 |
|---|
-Sugars , 2 types, 8 molecules 
| #3: Polysaccharide | Source method: isolated from a genetically manipulated source #4: Sugar | ChemComp-NAG / |
|---|
-Non-polymers , 3 types, 72 molecules 



| #5: Chemical | ChemComp-NO3 / #6: Chemical | ChemComp-BCN / | #7: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | N |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.36 Å3/Da / Density % sol: 47.9 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion / pH: 8.5 Details: 12.5% (v/v) MPD, 12.5% (v/v) PEG 1000, 12.5% (w/v) PEG 3350, 0.1 M Tris base/Bicine pH 8.5, 0.09 M NPS (nitrate, phosphate, sulfate; MD2-100-72, Molecular Dimensions). |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID30B / Wavelength: 1.7712 Å / Beamline: ID30B / Wavelength: 1.7712 Å |
| Detector | Type: DECTRIS PILATUS3 6M / Detector: PIXEL / Date: Jun 22, 2021 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.7712 Å / Relative weight: 1 |
| Reflection | Resolution: 2.9→38.02 Å / Num. obs: 40766 / % possible obs: 99.1 % / Redundancy: 3.2 % / CC1/2: 0.96 / CC star: 0.99 / Rmerge(I) obs: 0.233 / Rpim(I) all: 0.154 / Rrim(I) all: 0.28 / Net I/σ(I): 4.9 |
| Reflection shell | Resolution: 2.9→2.986 Å / Redundancy: 2.9 % / Rmerge(I) obs: 1.11 / Mean I/σ(I) obs: 1 / Num. unique obs: 3372 / CC1/2: 0.32 / CC star: 0.7 / Rpim(I) all: 0.765 / Rrim(I) all: 1.353 / % possible all: 99.6 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: AlphaFold model Resolution: 2.9→38 Å / Cross valid method: FREE R-VALUE / Phase error: 21.6235 Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 48.51 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.9→38 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller






 PDBj
PDBj


