[English] 日本語
 Yorodumi
Yorodumi- PDB-7zxa: Crystal structure of human cathepsin L with covalently bound alox... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7zxa | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure of human cathepsin L with covalently bound aloxistatin (E-64D) | |||||||||||||||||||||
 Components Components | Cathepsin L | |||||||||||||||||||||
 Keywords Keywords | HYDROLASE / cystein protease / drug target / lysosome / virus cell entry | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationenkephalin processing / cathepsin L / CD4-positive, alpha-beta T cell lineage commitment / macrophage apoptotic process / chromaffin granule / elastin catabolic process / antigen processing and presentation of peptide antigen / RUNX1 regulates transcription of genes involved in differentiation of keratinocytes / endolysosome lumen / cellular response to thyroid hormone stimulus ...enkephalin processing / cathepsin L / CD4-positive, alpha-beta T cell lineage commitment / macrophage apoptotic process / chromaffin granule / elastin catabolic process / antigen processing and presentation of peptide antigen / RUNX1 regulates transcription of genes involved in differentiation of keratinocytes / endolysosome lumen / cellular response to thyroid hormone stimulus / Trafficking and processing of endosomal TLR / proteoglycan binding / zymogen activation / Assembly of collagen fibrils and other multimeric structures / antigen processing and presentation / Collagen degradation / protein autoprocessing / collagen catabolic process / fibronectin binding / serpin family protein binding / collagen binding / Degradation of the extracellular matrix / Attachment and Entry / receptor-mediated endocytosis of virus by host cell / multivesicular body / endocytic vesicle lumen / MHC class II antigen presentation / cysteine-type peptidase activity / lysosomal lumen / : / Endosomal/Vacuolar pathway / antigen processing and presentation of exogenous peptide antigen via MHC class II / : / histone binding / adaptive immune response / Attachment and Entry / lysosome / apical plasma membrane / fusion of virus membrane with host plasma membrane / cysteine-type endopeptidase activity / intracellular membrane-bounded organelle / fusion of virus membrane with host endosome membrane / symbiont entry into host cell / Golgi apparatus / proteolysis / extracellular space / extracellular exosome / extracellular region / nucleus / plasma membrane Similarity search - Function | |||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.6 Å MOLECULAR REPLACEMENT / Resolution: 1.6 Å | |||||||||||||||||||||
 Authors Authors | Falke, S. / Lieske, J. / Guenther, S. / Reinke, P.Y.A. / Ewert, W. / Loboda, J. / Karnicar, K. / Usenik, A. / Lindic, N. / Sekirnik, A. ...Falke, S. / Lieske, J. / Guenther, S. / Reinke, P.Y.A. / Ewert, W. / Loboda, J. / Karnicar, K. / Usenik, A. / Lindic, N. / Sekirnik, A. / Chapman, H.N. / Hinrichs, W. / Turk, D. / Meents, A. | |||||||||||||||||||||
| Funding support |  Germany, Germany,  Slovenia, 6items Slovenia, 6items
| |||||||||||||||||||||
 Citation Citation |  Journal: J.Med.Chem. / Year: 2024 Journal: J.Med.Chem. / Year: 2024Title: Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors. Authors: Falke, S. / Lieske, J. / Herrmann, A. / Loboda, J. / Karnicar, K. / Gunther, S. / Reinke, P.Y.A. / Ewert, W. / Usenik, A. / Lindic, N. / Sekirnik, A. / Dretnik, K. / Tsuge, H. / Turk, V. / ...Authors: Falke, S. / Lieske, J. / Herrmann, A. / Loboda, J. / Karnicar, K. / Gunther, S. / Reinke, P.Y.A. / Ewert, W. / Usenik, A. / Lindic, N. / Sekirnik, A. / Dretnik, K. / Tsuge, H. / Turk, V. / Chapman, H.N. / Hinrichs, W. / Ebert, G. / Turk, D. / Meents, A. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7zxa.cif.gz 7zxa.cif.gz | 638.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7zxa.ent.gz pdb7zxa.ent.gz | 442.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7zxa.json.gz 7zxa.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/zx/7zxa https://data.pdbj.org/pub/pdb/validation_reports/zx/7zxa ftp://data.pdbj.org/pub/pdb/validation_reports/zx/7zxa ftp://data.pdbj.org/pub/pdb/validation_reports/zx/7zxa | HTTPS FTP |
|---|
-Related structure data
| Related structure data | 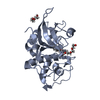 7qkdC  7zs7C  7zvfC  8a4uC  8a4vC  8a4wC  8a4xC  8a5bC  8ahvC  8b4fC  8c77C  8ofaC  8ozaC  8prxC  8qkbC  7z58S C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||
| 2 | 
| ||||||||||||
| 3 | 
| ||||||||||||
| 4 | 
| ||||||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 4 molecules ABCD
| #1: Protein | Mass: 24161.676 Da / Num. of mol.: 4 / Mutation: T110A Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: CTSL, CTSL1 / Production host: Homo sapiens (human) / Gene: CTSL, CTSL1 / Production host:  Komagataella phaffii GS115 (fungus) / References: UniProt: P07711 Komagataella phaffii GS115 (fungus) / References: UniProt: P07711 |
|---|
-Non-polymers , 9 types, 677 molecules 
















| #2: Chemical | ChemComp-E6D / #3: Chemical | ChemComp-1PE / | #4: Chemical | #5: Chemical | ChemComp-DMS / #6: Chemical | ChemComp-PEG / #7: Chemical | ChemComp-PGE / #8: Chemical | ChemComp-EDO / #9: Chemical | ChemComp-P6G / | #10: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.11 Å3/Da / Density % sol: 41.61 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 4 Details: Mature cathepsin L at a concentration of 7 mg/ml was equilibrated against 27% w/v PEG 8000, 1 mM TCEP and 0.1 M sodium acetate at pH 4.0. Crystals, which grew at 293 K to final size after ...Details: Mature cathepsin L at a concentration of 7 mg/ml was equilibrated against 27% w/v PEG 8000, 1 mM TCEP and 0.1 M sodium acetate at pH 4.0. Crystals, which grew at 293 K to final size after approximately 3 days, were transferred to a compound soaking solution containing 22% w/v PEG 8000, 1 mM TCEP and 0.1 M sodium acetate at pH 4.0 as well as 5% v/v DMSO and 10% v/v PEG 400. |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  PETRA III, DESY PETRA III, DESY  / Beamline: P11 / Wavelength: 1.033 Å / Beamline: P11 / Wavelength: 1.033 Å |
| Detector | Type: DECTRIS EIGER2 X 16M / Detector: PIXEL / Date: Apr 9, 2022 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.033 Å / Relative weight: 1 |
| Reflection | Resolution: 1.6→49.38 Å / Num. obs: 97574 / % possible obs: 93.63 % / Redundancy: 11.2 % / Biso Wilson estimate: 21.32 Å2 / CC1/2: 0.999 / CC star: 1 / Rmerge(I) obs: 0.1368 / Rpim(I) all: 0.0425 / Rrim(I) all: 0.1433 / Net I/σ(I): 11.28 |
| Reflection shell | Resolution: 1.6→1.657 Å / Redundancy: 11.1 % / Rmerge(I) obs: 2.232 / Mean I/σ(I) obs: 1.03 / Num. unique obs: 9570 / CC1/2: 0.52 / CC star: 0.827 / Rpim(I) all: 0.6974 / Rrim(I) all: 2.34 / % possible all: 91.89 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 7Z58 Resolution: 1.6→49.38 Å / SU ML: 0.1719 / Cross valid method: FREE R-VALUE / σ(F): 1.96 / Phase error: 22.1276 Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 31.33 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.6→49.38 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group | Refine-ID: X-RAY DIFFRACTION
|
 Movie
Movie Controller
Controller


 PDBj
PDBj




















