[English] 日本語
 Yorodumi
Yorodumi- PDB-7vyq: Short chain dehydrogenase (SCR) cryoEM structure with NADP and et... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7vyq | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Short chain dehydrogenase (SCR) cryoEM structure with NADP and ethyl 4-chloroacetoacetate | |||||||||
 Components Components | Carbonyl Reductase | |||||||||
 Keywords Keywords | OXIDOREDUCTASE / Rossman fold / tetramer / tag-free / wild type with NADPH | |||||||||
| Function / homology |  Function and homology information Function and homology informationmannitol 2-dehydrogenase (NADP+) activity / oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor / small molecule metabolic process / Oxidoreductases; Acting on the CH-OH group of donors; With NAD+ or NADP+ as acceptor / carbohydrate metabolic process Similarity search - Function | |||||||||
| Biological species |  Candida parapsilosis (yeast) Candida parapsilosis (yeast) | |||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.13 Å | |||||||||
 Authors Authors | Li, Y.H. / Zhang, R.Z. / Wang, C. / Forouhar, F. / Clarke, O. / Vorobiev, S. / Singh, S. / Montelione, G.T. / Szyperski, T. / Xu, Y. / Hunt, J.F. | |||||||||
| Funding support |  China, 2items China, 2items
| |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2022 Journal: EMBO J / Year: 2022Title: Oligomeric interactions maintain active-site structure in a noncooperative enzyme family. Authors: Yaohui Li / Rongzhen Zhang / Chi Wang / Farhad Forouhar / Oliver B Clarke / Sergey Vorobiev / Shikha Singh / Gaetano T Montelione / Thomas Szyperski / Yan Xu / John F Hunt /   Abstract: The evolutionary benefit accounting for widespread conservation of oligomeric structures in proteins lacking evidence of intersubunit cooperativity remains unclear. Here, crystal and cryo-EM ...The evolutionary benefit accounting for widespread conservation of oligomeric structures in proteins lacking evidence of intersubunit cooperativity remains unclear. Here, crystal and cryo-EM structures, and enzymological data, demonstrate that a conserved tetramer interface maintains the active-site structure in one such class of proteins, the short-chain dehydrogenase/reductase (SDR) superfamily. Phylogenetic comparisons support a significantly longer polypeptide being required to maintain an equivalent active-site structure in the context of a single subunit. Oligomerization therefore enhances evolutionary fitness by reducing the metabolic cost of enzyme biosynthesis. The large surface area of the structure-stabilizing oligomeric interface yields a synergistic gain in fitness by increasing tolerance to activity-enhancing yet destabilizing mutations. We demonstrate that two paralogous SDR superfamily enzymes with different specificities can form mixed heterotetramers that combine their individual enzymological properties. This suggests that oligomerization can also diversify the functions generated by a given metabolic investment, enhancing the fitness advantage provided by this architectural strategy. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7vyq.cif.gz 7vyq.cif.gz | 404.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7vyq.ent.gz pdb7vyq.ent.gz | 337.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7vyq.json.gz 7vyq.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/vy/7vyq https://data.pdbj.org/pub/pdb/validation_reports/vy/7vyq ftp://data.pdbj.org/pub/pdb/validation_reports/vy/7vyq ftp://data.pdbj.org/pub/pdb/validation_reports/vy/7vyq | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  32211MC  7dldC  7dllC  7dlmC  7dmgC  7dn1C M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
| EM raw data |  EMPIAR-10872 (Title: Oligomeric interactions maintain active-site structure in a non-cooperative enzyme family EMPIAR-10872 (Title: Oligomeric interactions maintain active-site structure in a non-cooperative enzyme familyData size: 902.0 Data #1: Native-SCR-tetramer-frames [micrographs - multiframe]) |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
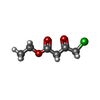
| #1: Protein | Mass: 30204.873 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Candida parapsilosis (yeast) / Gene: DQ675534 / Production host: Candida parapsilosis (yeast) / Gene: DQ675534 / Production host:  References: UniProt: B2KJ46, Oxidoreductases; Acting on the CH-OH group of donors; With NAD+ or NADP+ as acceptor #2: Chemical | ChemComp-NAP / #3: Chemical | ChemComp-83I / Has ligand of interest | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Ternary complex of SCR with NADP and ethyl 4-chloroacetoacetate Type: COMPLEX / Entity ID: #1 / Source: RECOMBINANT |
|---|---|
| Source (natural) | Organism:  Candida parapsilosis (yeast) Candida parapsilosis (yeast) |
| Source (recombinant) | Organism:  |
| Buffer solution | pH: 6 |
| Specimen | Conc.: 3 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 105000 X / Nominal defocus max: 2170 nm / Nominal defocus min: 1370 nm |
| Image recording | Average exposure time: 2.5 sec. / Electron dose: 56.15 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Num. of grids imaged: 1 / Num. of real images: 2885 |
- Processing
Processing
| Software | Name: PHENIX / Version: dev_4376: / Classification: refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
| Symmetry | Point symmetry: D2 (2x2 fold dihedral) | ||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.13 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 104278 / Symmetry type: POINT | ||||||||||||||||||||||||
| Atomic model building | B value: 177 / Space: REAL | ||||||||||||||||||||||||
| Atomic model building | PDB-ID: 7DLM Accession code: 7DLM / Source name: PDB / Type: experimental model | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller




 PDBj
PDBj