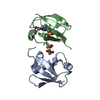
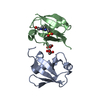
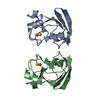
登録情報 データベース : PDB / ID : 7s6oタイトル The crystal structure of Lys48-linked di-ubiquitin (Ubiquitin) x 2 キーワード / / 機能・相同性 分子機能 ドメイン・相同性 構成要素
/ / / / / / / / / 生物種 Homo sapiens (ヒト)手法 / / / 解像度 : 1.25 Å データ登録者 Osipiuk, J. / Tesar, C. / Lanham, B.T. / Wydorski, P. / Fushman, D. / Joachimiak, L. / Joachimiak, A. 資金援助 組織 認可番号 国 Department of Energy (DOE, United States) DE-AC02-06CH11357
ジャーナル : Nat Commun / 年 : 2023タイトル : Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.著者 : Wydorski, P.M. / Osipiuk, J. / Lanham, B.T. / Tesar, C. / Endres, M. / Engle, E. / Jedrzejczak, R. / Mullapudi, V. / Michalska, K. / Fidelis, K. / Fushman, D. / Joachimiak, A. / Joachimiak, L.A. 履歴 登録 2021年9月14日 登録サイト / 処理サイト 改定 1.0 2021年9月22日 Provider / タイプ 改定 1.1 2021年12月15日 Group / Structure summaryカテゴリ / pdbx_audit_support / struct_keywordsItem 改定 2.0 2022年3月16日 Group Advisory / Atomic model ... Advisory / Atomic model / Author supporting evidence / Data collection / Database references / Derived calculations / Non-polymer description / Polymer sequence / Refinement description / Source and taxonomy / Structure summary カテゴリ atom_site / atom_site_anisotrop ... atom_site / atom_site_anisotrop / chem_comp / entity / entity_poly / entity_poly_seq / entity_src_gen / pdbx_audit_support / pdbx_contact_author / pdbx_database_related / pdbx_entity_nonpoly / pdbx_entry_details / pdbx_nonpoly_scheme / pdbx_poly_seq_scheme / pdbx_struct_assembly / pdbx_struct_assembly_gen / pdbx_struct_assembly_prop / pdbx_struct_sheet_hbond / pdbx_unobs_or_zero_occ_residues / pdbx_validate_close_contact / refine / refine_hist / refine_ls_restr / refine_ls_shell / reflns / reflns_shell / software / struct_asym / struct_conf / struct_ref_seq_dif / struct_sheet_range Item _chem_comp.formula / _chem_comp.formula_weight ... _chem_comp.formula / _chem_comp.formula_weight / _chem_comp.id / _chem_comp.mon_nstd_flag / _chem_comp.name / _chem_comp.type / _entity_poly.pdbx_seq_one_letter_code / _entity_poly.pdbx_seq_one_letter_code_can / _entity_poly_seq.entity_id / _entity_poly_seq.mon_id / _entity_poly_seq.num / _entity_src_gen.gene_src_common_name / _entity_src_gen.pdbx_end_seq_num / _pdbx_poly_seq_scheme.asym_id / _pdbx_poly_seq_scheme.auth_mon_id / _pdbx_poly_seq_scheme.auth_seq_num / _pdbx_poly_seq_scheme.entity_id / _pdbx_poly_seq_scheme.mon_id / _pdbx_poly_seq_scheme.ndb_seq_num / _pdbx_poly_seq_scheme.pdb_mon_id / _pdbx_poly_seq_scheme.pdb_seq_num / _pdbx_poly_seq_scheme.pdb_strand_id / _pdbx_poly_seq_scheme.seq_id / _pdbx_struct_assembly.details / _pdbx_struct_assembly.method_details / _pdbx_struct_assembly_gen.asym_id_list / _pdbx_struct_assembly_prop.value / _pdbx_struct_sheet_hbond.range_1_auth_comp_id / _pdbx_struct_sheet_hbond.range_1_auth_seq_id / _pdbx_struct_sheet_hbond.range_1_label_comp_id / _pdbx_struct_sheet_hbond.range_1_label_seq_id / _pdbx_struct_sheet_hbond.range_2_auth_comp_id / _pdbx_struct_sheet_hbond.range_2_auth_seq_id / _pdbx_struct_sheet_hbond.range_2_label_comp_id / _pdbx_struct_sheet_hbond.range_2_label_seq_id / _pdbx_unobs_or_zero_occ_residues.auth_asym_id / _pdbx_unobs_or_zero_occ_residues.auth_comp_id / _pdbx_unobs_or_zero_occ_residues.auth_seq_id / _pdbx_unobs_or_zero_occ_residues.label_asym_id / _pdbx_unobs_or_zero_occ_residues.label_comp_id / _pdbx_unobs_or_zero_occ_residues.label_seq_id / _refine.B_iso_max / _refine.B_iso_mean / _refine.B_iso_min / _refine.aniso_B[1][1] / _refine.aniso_B[1][3] / _refine.aniso_B[2][2] / _refine.aniso_B[2][3] / _refine.aniso_B[3][3] / _refine.correlation_coeff_Fo_to_Fc / _refine.correlation_coeff_Fo_to_Fc_free / _refine.ls_R_factor_R_free / _refine.ls_R_factor_R_work / _refine.ls_R_factor_obs / _refine.overall_SU_B / _refine.overall_SU_ML / _refine.pdbx_overall_ESU_R / _refine.pdbx_overall_ESU_R_Free / _refine_hist.number_atoms_solvent / _refine_hist.number_atoms_total / _refine_hist.pdbx_B_iso_mean_ligand / _refine_hist.pdbx_B_iso_mean_solvent / _refine_hist.pdbx_number_atoms_ligand / _refine_ls_restr.dev_ideal / _refine_ls_restr.dev_ideal_target / _refine_ls_restr.number / _refine_ls_shell.R_factor_R_free / _refine_ls_shell.R_factor_R_work / _reflns.d_resolution_low / _reflns.pdbx_CC_half / _reflns.pdbx_CC_star / _reflns.pdbx_netI_over_av_sigmaI / _reflns.pdbx_number_measured_all / _reflns_shell.d_res_low / _reflns_shell.meanI_over_sigI_obs / _reflns_shell.pdbx_CC_star / _software.classification / _software.name / _software.version / _struct_conf.beg_auth_comp_id / _struct_conf.beg_auth_seq_id / _struct_conf.beg_label_comp_id / _struct_conf.beg_label_seq_id / _struct_conf.pdbx_PDB_helix_length / _struct_ref_seq_dif.db_mon_id / _struct_ref_seq_dif.details / _struct_ref_seq_dif.mon_id / _struct_ref_seq_dif.pdbx_auth_seq_num / _struct_ref_seq_dif.pdbx_seq_db_seq_num / _struct_ref_seq_dif.seq_num / _struct_sheet_range.beg_auth_comp_id / _struct_sheet_range.beg_label_comp_id 解説 詳細: The previous deposition based on small differences in electron density maps and had wrong assignment of chains due to invisibility of linker between molecules. We were informed about similar ... 詳細 Provider / タイプ 改定 2.1 2023年5月31日 Group / カテゴリ / citation_authorItem _citation.country / _citation.journal_abbrev ... _citation.country / _citation.journal_abbrev / _citation.journal_id_CSD / _citation.journal_id_ISSN / _citation.journal_volume / _citation.page_first / _citation.page_last / _citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed / _citation.title / _citation.year 改定 2.2 2023年10月25日 Group / Refinement descriptionカテゴリ / chem_comp_bond / pdbx_initial_refinement_model
すべて表示 表示を減らす
 データを開く
データを開く 基本情報
基本情報 要素
要素 キーワード
キーワード 機能・相同性情報
機能・相同性情報 Homo sapiens (ヒト)
Homo sapiens (ヒト) X線回折 /
X線回折 /  シンクロトロン /
シンクロトロン /  分子置換 / 解像度: 1.25 Å
分子置換 / 解像度: 1.25 Å  データ登録者
データ登録者 米国, 1件
米国, 1件  引用
引用 ジャーナル: Nat Commun / 年: 2023
ジャーナル: Nat Commun / 年: 2023 構造の表示
構造の表示 Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク ダウンロード
ダウンロード 7s6o.cif.gz
7s6o.cif.gz PDBx/mmCIF形式
PDBx/mmCIF形式 pdb7s6o.ent.gz
pdb7s6o.ent.gz PDB形式
PDB形式 7s6o.json.gz
7s6o.json.gz PDBx/mmJSON形式
PDBx/mmJSON形式 その他のダウンロード
その他のダウンロード 7s6o_validation.pdf.gz
7s6o_validation.pdf.gz wwPDB検証レポート
wwPDB検証レポート 7s6o_full_validation.pdf.gz
7s6o_full_validation.pdf.gz 7s6o_validation.xml.gz
7s6o_validation.xml.gz 7s6o_validation.cif.gz
7s6o_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/s6/7s6o
https://data.pdbj.org/pub/pdb/validation_reports/s6/7s6o ftp://data.pdbj.org/pub/pdb/validation_reports/s6/7s6o
ftp://data.pdbj.org/pub/pdb/validation_reports/s6/7s6o リンク
リンク 集合体
集合体
 要素
要素 Homo sapiens (ヒト) / 遺伝子: UBB / 発現宿主:
Homo sapiens (ヒト) / 遺伝子: UBB / 発現宿主: 
 Homo sapiens (ヒト) / 遺伝子: UBB / 発現宿主:
Homo sapiens (ヒト) / 遺伝子: UBB / 発現宿主: 
 X線回折 / 使用した結晶の数: 1
X線回折 / 使用した結晶の数: 1  試料調製
試料調製 シンクロトロン / サイト:
シンクロトロン / サイト:  APS
APS  / ビームライン: 19-ID / 波長: 0.9792 Å
/ ビームライン: 19-ID / 波長: 0.9792 Å 解析
解析 分子置換
分子置換 ムービー
ムービー コントローラー
コントローラー
















 PDBj
PDBj




