[English] 日本語
 Yorodumi
Yorodumi- PDB-7nso: Structure of ErmDL-Erythromycin-stalled 70S E. coli ribosomal com... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7nso | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of ErmDL-Erythromycin-stalled 70S E. coli ribosomal complex with P-tRNA | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | RIBOSOME / Antibiotic / ErmDL / Erythromycin / Macrolide / Stalling | |||||||||
| Function / homology |  Function and homology information Function and homology informationtranscriptional attenuation / endoribonuclease inhibitor activity / RNA-binding transcription regulator activity / negative regulation of cytoplasmic translation / DnaA-L2 complex / translation repressor activity / negative regulation of DNA-templated DNA replication initiation / assembly of large subunit precursor of preribosome / cytosolic ribosome assembly / ribosome assembly ...transcriptional attenuation / endoribonuclease inhibitor activity / RNA-binding transcription regulator activity / negative regulation of cytoplasmic translation / DnaA-L2 complex / translation repressor activity / negative regulation of DNA-templated DNA replication initiation / assembly of large subunit precursor of preribosome / cytosolic ribosome assembly / ribosome assembly / DNA-templated transcription termination / response to radiation / mRNA 5'-UTR binding / large ribosomal subunit / transferase activity / ribosomal small subunit assembly / ribosomal large subunit assembly / 5S rRNA binding / small ribosomal subunit / small ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / large ribosomal subunit rRNA binding / cytosolic large ribosomal subunit / cytoplasmic translation / tRNA binding / negative regulation of translation / rRNA binding / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / response to antibiotic / negative regulation of DNA-templated transcription / mRNA binding / DNA binding / RNA binding / zinc ion binding / membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Beckert, B. / Wilson, D.N. | |||||||||
| Funding support |  Germany, 2items Germany, 2items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Structural and mechanistic basis for translation inhibition by macrolide and ketolide antibiotics. Authors: Bertrand Beckert / Elodie C Leroy / Shanmugapriya Sothiselvam / Lars V Bock / Maxim S Svetlov / Michael Graf / Stefan Arenz / Maha Abdelshahid / Britta Seip / Helmut Grubmüller / Alexander ...Authors: Bertrand Beckert / Elodie C Leroy / Shanmugapriya Sothiselvam / Lars V Bock / Maxim S Svetlov / Michael Graf / Stefan Arenz / Maha Abdelshahid / Britta Seip / Helmut Grubmüller / Alexander S Mankin / C Axel Innis / Nora Vázquez-Laslop / Daniel N Wilson /    Abstract: Macrolides and ketolides comprise a family of clinically important antibiotics that inhibit protein synthesis by binding within the exit tunnel of the bacterial ribosome. While these antibiotics are ...Macrolides and ketolides comprise a family of clinically important antibiotics that inhibit protein synthesis by binding within the exit tunnel of the bacterial ribosome. While these antibiotics are known to interrupt translation at specific sequence motifs, with ketolides predominantly stalling at Arg/Lys-X-Arg/Lys motifs and macrolides displaying a broader specificity, a structural basis for their context-specific action has been lacking. Here, we present structures of ribosomes arrested during the synthesis of an Arg-Leu-Arg sequence by the macrolide erythromycin (ERY) and the ketolide telithromycin (TEL). Together with deep mutagenesis and molecular dynamics simulations, the structures reveal how ERY and TEL interplay with the Arg-Leu-Arg motif to induce translational arrest and illuminate the basis for the less stringent sequence-specific action of ERY over TEL. Because programmed stalling at the Arg/Lys-X-Arg/Lys motifs is used to activate expression of antibiotic resistance genes, our study also provides important insights for future development of improved macrolide antibiotics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7nso.cif.gz 7nso.cif.gz | 3.1 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7nso.ent.gz pdb7nso.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  7nso.json.gz 7nso.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ns/7nso https://data.pdbj.org/pub/pdb/validation_reports/ns/7nso ftp://data.pdbj.org/pub/pdb/validation_reports/ns/7nso ftp://data.pdbj.org/pub/pdb/validation_reports/ns/7nso | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  12573MC  7nspC  7nsqC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-RNA chain , 5 types, 5 molecules AB68a
| #1: RNA chain | Mass: 941306.188 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #2: RNA chain | Mass: 38790.090 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #32: RNA chain | Mass: 872.556 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #34: RNA chain | Mass: 28094.645 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #35: RNA chain | Mass: 499054.625 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
+50S ribosomal protein ... , 29 types, 29 molecules CDEFGHJKLMNOPQRSTUVWXYZ012345
-30S ribosomal protein ... , 20 types, 20 molecules bcdefghijklmnopqrstu
| #36: Protein | Mass: 24971.764 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #37: Protein | Mass: 23078.785 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #38: Protein | Mass: 23383.002 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #39: Protein | Mass: 16532.088 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #40: Protein | Mass: 11582.294 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #41: Protein | Mass: 16861.523 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #42: Protein | Mass: 14015.361 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #43: Protein | Mass: 14554.882 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #44: Protein | Mass: 11196.988 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #45: Protein | Mass: 12487.200 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #46: Protein | Mass: 13636.961 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #47: Protein | Mass: 12625.753 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #48: Protein | Mass: 11475.364 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #49: Protein | Mass: 10159.621 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #50: Protein | Mass: 9207.572 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #51: Protein | Mass: 9263.946 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #52: Protein | Mass: 7734.896 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #53: Protein | Mass: 9421.018 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #54: Protein | Mass: 9577.268 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #55: Protein | Mass: 8392.844 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
-Protein/peptide / Non-polymers , 2 types, 2 molecules 7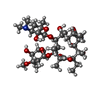

| #33: Protein/peptide | Mass: 877.087 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #56: Chemical | ChemComp-ERY / |
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Structure of ErmDL-Erythromycin-stalled 70S E. coli ribosomal complex with P-tRNA Type: RIBOSOME / Entity ID: #1-#55 / Source: NATURAL |
|---|---|
| Molecular weight | Experimental value: YES |
| Source (natural) | Organism:  |
| Buffer solution | pH: 7.5 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Specimen support | Grid material: COPPER / Grid mesh size: 300 divisions/in. / Grid type: Quantifoil R3/3 |
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE-PROPANE / Humidity: 100 % |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: SPOT SCAN FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: SPOT SCAN |
| Electron lens | Mode: BRIGHT FIELD / Cs: 2.7 mm / Alignment procedure: COMA FREE |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Average exposure time: 35 sec. / Electron dose: 1 e/Å2 / Detector mode: COUNTING / Film or detector model: FEI FALCON II (4k x 4k) |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 288013 | ||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry: C1 (asymmetric) | ||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 2.9 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 172175 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||
| Atomic model building | Protocol: RIGID BODY FIT / Space: REAL |
 Movie
Movie Controller
Controller
























 PDBj
PDBj































