| 1 | 300 uM [U-13C; U-15N] Tpt1, 20 mM HEPES, 200 mM sodium chloride, 2 mM DTT, 2 mM [U-100% 2H] EDTA, 5 % v/v [U-100% 2H] glycerol, 0.5 mM AEBSF protease inhibitor, 0.04 % v/v sodium azide, 50 uM DSS, 95% H2O/5% D2O | 15N-13C_sample1 | 95% H2O/5% D2O | solution| 2 | 360 uM [U-13C; U-15N] Tpt1, 20 mM HEPES, 200 mM sodium chloride, 2 mM DTT, 2 mM [U-100% 2H] EDTA, 5 % v/v [U-100% 2H] glycerol, 0.5 mM AEBSF protease inhibitor, 0.04 % v/v sodium azide, 50 uM DSS, 95% H2O/5% D2O | 15N-13C_sample2 | 95% H2O/5% D2O | solution| 3 | 300 uM [U-13C; U-15N] Tpt1, 20 mM HEPES, 200 mM sodium chloride, 2 mM DTT, 2 mM [U-100% 2H] EDTA, 5 % v/v [U-100% 2H] glycerol, 1 mM AEBSF protease inhibitor, 0.05 % w/v sodium azide, 50 uM DSS, 95% H2O/5% D2O | 15N-13C_sample3 | 95% H2O/5% D2O | solution| 4 | 400 uM [U-13C; U-15N] Tpt1, 20 mM HEPES, 200 mM sodium chloride, 2 mM DTT, 2 mM [U-100% 2H] EDTA, 5 % v/v [U-100% 2H] glycerol, 0.5 mM AEBSF protease inhibitor, 0.05 % w/v sodium azide, 50 mM DSS, 100% D2O | 15N-13C_sample4_d2o | 100% D2O | solution| 7 | 420 uM [U-13C; U-15N] Tpt1, 20 mM HEPES, 200 mM sodium chloride, 2 mM DTT, 2 mM [U-100% 2H] EDTA, 5 % v/v [U-100% 2H] glycerol, 0.5 mM AEBSF protease inhibitor, 0.04 % w/v sodium azide, 50 mM DSS, 95% H2O/5% D2O | 15N-13C_sample7 | 95% H2O/5% D2O | solution| 8 | 380 uM [U-13C; U-15N] Tpt1, 20 mM HEPES, 200 mM sodium chloride, 2 mM DTT, 2 mM [U-100% 2H] EDTA, 5 % v/v [U-100% 2H] glycerol, 0.5 mM AEBSF protease inhibitor, 0.04 % w/v sodium azide, 50 mM DSS, 95% H2O/5% D2O | 15N-13C_sample8 | 95% H2O/5% D2O | solution| 9 | 400 uM [U-13C; U-15N] Tpt1, 20 mM HEPES, 200 mM sodium chloride, 2 mM DTT, 2 mM [U-100% 2H] EDTA, 5 % v/v [U-100% 2H] glycerol, 0.5 mM AEBSF protease inhibitor, 0.04 % w/v sodium azide, 50 mM DSS, 100% D2O | 15N-13C_sample9_d2o | 100% D2O | solution| 5 | 400 uM [U-2H; U-15N];[13CD1,1H3]ILE;[13CD1,1H3;13CD2,1H3]LEU;[13CG1,1H3;13CG2,1H3]VAL;[13CE1,1H3]MET;[13CG2,1H3]THR Tpt1, 20 mM HEPES, 200 mM sodium chloride, 2 mM DTT, 2 mM [U-100% 2H] EDTA, 5 % v/v [U-100% 2H] glycerol, 0.5 mM AEBSF protease inhibitor, 0.05 % w/v sodium azide, 50 uM DSS, 100% D2O | 2H-15N_ILVMT_sample5_d2O | 100% D2O| Uniformly [2H-15N] sample with [13C-1H] labeling on methyl groups for ILE [d1 only] VAL (g1 and g2), Leu (d1 and d2) and THR (g2). | solution| 6 | 350 uM [U-2H; U-15N];[13CD1,1H3]ILE;[13CD1,1H3;13CD2,1H3]LEU;[13CG1,1H3;13CG2,1H3]VAL;[13CE1,1H3]MET;[13CG2,1H3]THR Tpt1, 20 mM HEPES, 200 mM sodium chloride, 2 mM DTT, 2 mM [U-100% 2H] EDTA, 5 % v/v [U-100% 2H] glycerol, 0.5 mM AEBSF protease inhibitor, 0.05 % w/v sodium azide, 50 uM DSS, 100% D2O | 2H-15N_ILVMT_sample6_d2O | 100% D2O| Uniformly [2H-15N] sample with [13C-1H] labeling on methyl groups for ILE [d1 only] VAL (g1 and g2), Leu (d1 and d2) and THR (g2). | | | | | | | | | | | | | | | | | |  Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Runella slithyformis (bacteria)
Runella slithyformis (bacteria) Authors
Authors United States, 1items
United States, 1items  Citation
Citation Journal: Nucleic Acids Res. / Year: 2021
Journal: Nucleic Acids Res. / Year: 2021 Structure visualization
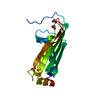
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 7kw8.cif.gz
7kw8.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb7kw8.ent.gz
pdb7kw8.ent.gz PDB format
PDB format 7kw8.json.gz
7kw8.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/kw/7kw8
https://data.pdbj.org/pub/pdb/validation_reports/kw/7kw8 ftp://data.pdbj.org/pub/pdb/validation_reports/kw/7kw8
ftp://data.pdbj.org/pub/pdb/validation_reports/kw/7kw8
 Links
Links Assembly
Assembly
 Components
Components Runella slithyformis (bacteria) / Production host:
Runella slithyformis (bacteria) / Production host: 
 Sample preparation
Sample preparation Movie
Movie Controller
Controller











 PDBj
PDBj HNCA
HNCA