[English] 日本語
 Yorodumi
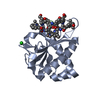
Yorodumi- PDB-7k0a: Puromycin N-acetyltransferase in complex with acetylated puromyci... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7k0a | ||||||
|---|---|---|---|---|---|---|---|
| Title | Puromycin N-acetyltransferase in complex with acetylated puromycin and CoA | ||||||
 Components Components | Puromycin N-acetyltransferase | ||||||
 Keywords Keywords | TRANSFERASE / Enzyme acetyletransferase complex selection marker / ANTIBIOTIC | ||||||
| Function / homology |  Function and homology information Function and homology informationTransferases; Acyltransferases / acyltransferase activity, transferring groups other than amino-acyl groups / response to antibiotic Similarity search - Function | ||||||
| Biological species |  Streptomyces alboniger (bacteria) Streptomyces alboniger (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2 Å MOLECULAR REPLACEMENT / Resolution: 2 Å | ||||||
 Authors Authors | Peat, T.S. / Caputo, A.T. / Newman, J. / Adams, T.E. | ||||||
 Citation Citation |  Journal: Sci Rep / Year: 2021 Journal: Sci Rep / Year: 2021Title: Structure-guided selection of puromycin N-acetyltransferase mutants with enhanced selection stringency for deriving mammalian cell lines expressing recombinant proteins. Authors: Caputo, A.T. / Eder, O.M. / Bereznakova, H. / Pothuis, H. / Ardevol, A. / Newman, J. / Nuttall, S. / Peat, T.S. / Adams, T.E. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7k0a.cif.gz 7k0a.cif.gz | 106.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7k0a.ent.gz pdb7k0a.ent.gz | 77 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7k0a.json.gz 7k0a.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/k0/7k0a https://data.pdbj.org/pub/pdb/validation_reports/k0/7k0a ftp://data.pdbj.org/pub/pdb/validation_reports/k0/7k0a ftp://data.pdbj.org/pub/pdb/validation_reports/k0/7k0a | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  7k09SC S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| |||||||||
| 2 | 
| |||||||||
| Unit cell |
| |||||||||
| Components on special symmetry positions |
| |||||||||
| Noncrystallographic symmetry (NCS) | NCS domain: (Details: Chains A B) |
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein | Mass: 22581.490 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Streptomyces alboniger (bacteria) / Gene: pac / Production host: Streptomyces alboniger (bacteria) / Gene: pac / Production host:  |
|---|
-Non-polymers , 6 types, 288 molecules 










| #2: Chemical | | #3: Chemical | #4: Chemical | #5: Chemical | ChemComp-SO4 / #6: Chemical | #7: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.79 Å3/Da / Density % sol: 55.94 % |
|---|---|
| Crystal grow | Temperature: 281 K / Method: vapor diffusion, sitting drop / pH: 8.26 Details: 9.9% (w/v) 1,6-hexanediol, 1.59 M ammonium sulfate, 7.4 % (v/v) polyethylene glycol 400, 0.1 M HEPES pH 8.26, 0.5 n-hexyl-b-D-glucopyranoside |
-Data collection
| Diffraction |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source |
| ||||||||||||||||||||
| Detector |
| ||||||||||||||||||||
| Radiation |
| ||||||||||||||||||||
| Radiation wavelength |
| ||||||||||||||||||||
| Reflection | Resolution: 2→70.75 Å / Num. obs: 33396 / % possible obs: 99.9 % / Redundancy: 17.2 % / CC1/2: 0.989 / Rmerge(I) obs: 0.376 / Rpim(I) all: 0.136 / Rrim(I) all: 0.4 / Χ2: 1.02 / Net I/σ(I): 7.7 | ||||||||||||||||||||
| Reflection shell | Resolution: 2→2.05 Å / Redundancy: 15.2 % / Mean I/σ(I) obs: 2.3 / Num. unique obs: 2460 / CC1/2: 0.857 / Rpim(I) all: 0.608 / % possible all: 99.4 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 7K09 Resolution: 2→42.488 Å / Cor.coef. Fo:Fc: 0.92 / Cor.coef. Fo:Fc free: 0.895 / SU B: 4.723 / SU ML: 0.126 / Cross valid method: FREE R-VALUE / ESU R: 0.187 / ESU R Free: 0.162 Details: Hydrogens have been added in their riding positions
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK BULK SOLVENT | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 16.23 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2→42.488 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj











