[English] 日本語
 Yorodumi
Yorodumi- PDB-7jpw: Rabbit Cav1.1 in the presence of 100 micromolar (R)-(+)-Bay K8644... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7jpw | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Rabbit Cav1.1 in the presence of 100 micromolar (R)-(+)-Bay K8644 in nanodiscs at 3.2 Angstrom resolution | |||||||||||||||||||||||||||||||||
 Components Components |
| |||||||||||||||||||||||||||||||||
 Keywords Keywords | TRANSPORT PROTEIN / rCav1.1 / Channels / Calcium Ion-Selective / drugs | |||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationhigh voltage-gated calcium channel activity / L-type voltage-gated calcium channel complex / positive regulation of muscle contraction / regulation of release of sequestered calcium ion into cytosol / regulation of calcium ion transmembrane transport via high voltage-gated calcium channel / cellular response to caffeine / calcium ion import across plasma membrane / voltage-gated calcium channel activity / release of sequestered calcium ion into cytosol / T-tubule ...high voltage-gated calcium channel activity / L-type voltage-gated calcium channel complex / positive regulation of muscle contraction / regulation of release of sequestered calcium ion into cytosol / regulation of calcium ion transmembrane transport via high voltage-gated calcium channel / cellular response to caffeine / calcium ion import across plasma membrane / voltage-gated calcium channel activity / release of sequestered calcium ion into cytosol / T-tubule / muscle contraction / calcium channel regulator activity / sarcolemma / calcium ion transmembrane transport / transmembrane transporter binding / calmodulin binding / metal ion binding / plasma membrane Similarity search - Function | |||||||||||||||||||||||||||||||||
| Biological species |  | |||||||||||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||||||||||||||||||||||||||
 Authors Authors | Yan, N. / Gao, S. | |||||||||||||||||||||||||||||||||
| Funding support |  United States, 1items United States, 1items
| |||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Angew Chem Int Ed Engl / Year: 2021 Journal: Angew Chem Int Ed Engl / Year: 2021Title: Structural Basis of the Modulation of the Voltage-Gated Calcium Ion Channel Ca 1.1 by Dihydropyridine Compounds*. Authors: Shuai Gao / Nieng Yan /  Abstract: 1,4-Dihydropyridines (DHP), the most commonly used antihypertensives, function by inhibiting the L-type voltage-gated Ca (Ca ) channels. DHP compounds exhibit chirality-specific antagonistic or ...1,4-Dihydropyridines (DHP), the most commonly used antihypertensives, function by inhibiting the L-type voltage-gated Ca (Ca ) channels. DHP compounds exhibit chirality-specific antagonistic or agonistic effects. The structure of rabbit Ca 1.1 bound to an achiral drug nifedipine reveals the general binding mode for DHP drugs, but the molecular basis for chiral specificity remained elusive. Herein, we report five cryo-EM structures of nanodisc-embedded Ca 1.1 in the presence of the bestselling drug amlodipine, a DHP antagonist (R)-(+)-Bay K8644, and a titration of its agonistic enantiomer (S)-(-)-Bay K8644 at resolutions of 2.9-3.4 Å. The amlodipine-bound structure reveals the molecular basis for the high efficacy of the drug. All structures with the addition of the Bay K8644 enantiomers exhibit similar inactivated conformations, suggesting that (S)-(-)-Bay K8644, when acting as an agonist, is insufficient to lock the activated state of the channel for a prolonged duration. | |||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7jpw.cif.gz 7jpw.cif.gz | 442.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7jpw.ent.gz pdb7jpw.ent.gz | 343 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7jpw.json.gz 7jpw.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/jp/7jpw https://data.pdbj.org/pub/pdb/validation_reports/jp/7jpw ftp://data.pdbj.org/pub/pdb/validation_reports/jp/7jpw ftp://data.pdbj.org/pub/pdb/validation_reports/jp/7jpw | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  22425MC  7jpkC  7jplC  7jpvC  7jpxC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 212240.594 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|
-Voltage-dependent calcium channel ... , 2 types, 2 molecules EF
| #2: Protein | Mass: 25082.254 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #3: Protein | Mass: 125082.906 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
-Non-polymers , 4 types, 12 molecules 


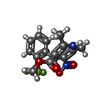



| #4: Chemical | ChemComp-CA / | ||||
|---|---|---|---|---|---|
| #5: Chemical | ChemComp-3PE / #6: Chemical | ChemComp-PC1 / | #7: Chemical | ChemComp-VFY / | |
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: rCav1.1-100R / Type: COMPLEX / Entity ID: #1-#3 / Source: NATURAL |
|---|---|
| Molecular weight | Experimental value: NO |
| Source (natural) | Organism:  |
| Buffer solution | pH: 8 |
| Specimen | Conc.: 5 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES / Details: rCav1.1-100R was in lipid nanodisc |
| Specimen support | Details: unspecified |
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 281 K / Details: blot for 5 seconds before plunging |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD |
| Image recording | Electron dose: 50 e/Å2 / Film or detector model: GATAN K2 SUMMIT (4k x 4k) |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.17rc2_3619: / Classification: refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software | Name: PHENIX / Category: model refinement | ||||||||||||||||||||||||
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.2 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 94695 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj










