[English] 日本語
 Yorodumi
Yorodumi- EMDB-22425: Rabbit Cav1.1 in the presence of 100 micromolar (R)-(+)-Bay K8644... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22425 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Rabbit Cav1.1 in the presence of 100 micromolar (R)-(+)-Bay K8644 in nanodiscs at 3.2 Angstrom resolution | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | rCav1.1 / Channels / Calcium Ion-Selective / TRANSPORT PROTEIN / drugs | |||||||||
| Function / homology |  Function and homology information Function and homology informationhigh voltage-gated calcium channel activity / L-type voltage-gated calcium channel complex / positive regulation of muscle contraction / regulation of release of sequestered calcium ion into cytosol / regulation of calcium ion transmembrane transport via high voltage-gated calcium channel / cellular response to caffeine / calcium ion import across plasma membrane / voltage-gated calcium channel activity / release of sequestered calcium ion into cytosol / T-tubule ...high voltage-gated calcium channel activity / L-type voltage-gated calcium channel complex / positive regulation of muscle contraction / regulation of release of sequestered calcium ion into cytosol / regulation of calcium ion transmembrane transport via high voltage-gated calcium channel / cellular response to caffeine / calcium ion import across plasma membrane / voltage-gated calcium channel activity / release of sequestered calcium ion into cytosol / T-tubule / muscle contraction / calcium channel regulator activity / sarcolemma / calcium ion transmembrane transport / transmembrane transporter binding / calmodulin binding / metal ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Yan N / Gao S | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Angew Chem Int Ed Engl / Year: 2021 Journal: Angew Chem Int Ed Engl / Year: 2021Title: Structural Basis of the Modulation of the Voltage-Gated Calcium Ion Channel Ca 1.1 by Dihydropyridine Compounds*. Authors: Shuai Gao / Nieng Yan /  Abstract: 1,4-Dihydropyridines (DHP), the most commonly used antihypertensives, function by inhibiting the L-type voltage-gated Ca (Ca ) channels. DHP compounds exhibit chirality-specific antagonistic or ...1,4-Dihydropyridines (DHP), the most commonly used antihypertensives, function by inhibiting the L-type voltage-gated Ca (Ca ) channels. DHP compounds exhibit chirality-specific antagonistic or agonistic effects. The structure of rabbit Ca 1.1 bound to an achiral drug nifedipine reveals the general binding mode for DHP drugs, but the molecular basis for chiral specificity remained elusive. Herein, we report five cryo-EM structures of nanodisc-embedded Ca 1.1 in the presence of the bestselling drug amlodipine, a DHP antagonist (R)-(+)-Bay K8644, and a titration of its agonistic enantiomer (S)-(-)-Bay K8644 at resolutions of 2.9-3.4 Å. The amlodipine-bound structure reveals the molecular basis for the high efficacy of the drug. All structures with the addition of the Bay K8644 enantiomers exhibit similar inactivated conformations, suggesting that (S)-(-)-Bay K8644, when acting as an agonist, is insufficient to lock the activated state of the channel for a prolonged duration. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22425.map.gz emd_22425.map.gz | 77 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22425-v30.xml emd-22425-v30.xml emd-22425.xml emd-22425.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_22425.png emd_22425.png | 43.1 KB | ||
| Filedesc metadata |  emd-22425.cif.gz emd-22425.cif.gz | 8.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22425 http://ftp.pdbj.org/pub/emdb/structures/EMD-22425 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22425 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22425 | HTTPS FTP |
-Related structure data
| Related structure data |  7jpwMC  7jpkC  7jplC  7jpvC  7jpxC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22425.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22425.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.114 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : rCav1.1-100R
| Entire | Name: rCav1.1-100R |
|---|---|
| Components |
|
-Supramolecule #1: rCav1.1-100R
| Supramolecule | Name: rCav1.1-100R / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Voltage-dependent L-type calcium channel subunit alpha-1S
| Macromolecule | Name: Voltage-dependent L-type calcium channel subunit alpha-1S type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 212.240594 KDa |
| Sequence | String: MEPSSPQDEG LRKKQPKKPL PEVLPRPPRA LFCLTLQNPL RKACISIVEW KPFETIILLT IFANCVALAV YLPMPEDDNN SLNLGLEKL EYFFLTVFSI EAAMKIIAYG FLFHQDAYLR SGWNVLDFII VFLGVFTAIL EQVNVIQSNT APMSSKGAGL D VKALRAFR ...String: MEPSSPQDEG LRKKQPKKPL PEVLPRPPRA LFCLTLQNPL RKACISIVEW KPFETIILLT IFANCVALAV YLPMPEDDNN SLNLGLEKL EYFFLTVFSI EAAMKIIAYG FLFHQDAYLR SGWNVLDFII VFLGVFTAIL EQVNVIQSNT APMSSKGAGL D VKALRAFR VLRPLRLVSG VPSLQVVLNS IFKAMLPLFH IALLVLFMVI IYAIIGLELF KGKMHKTCYY IGTDIVATVE NE KPSPCAR TGSGRPCTIN GSECRGGWPG PNHGITHFDN FGFSMLTVYQ CITMEGWTDV LYWVNDAIGN EWPWIYFVTL ILL GSFFIL NLVLGVLSGE FTKEREKAKS RGTFQKLREK QQLEEDLRGY MSWITQGEVM DVEDLREGKL SLEEGGSDTE SLYE IEGLN KIIQFIRHWR QWNRVFRWKC HDLVKSRVFY WLVILIVALN TLSIASEHHN QPLWLTHLQD IANRVLLSLF TIEML LKMY GLGLRQYFMS IFNRFDCFVV CSGILELLLV ESGAMTPLGI SVLRCIRLLR LFKITKYWTS LSNLVASLLN SIRSIA SLL LLLFLFIIIF ALLGMQLFGG RYDFEDTEVR RSNFDNFPQA LISVFQVLTG EDWNSVMYNG IMAYGGPSYP GVLVCIY FI ILFVCGNYIL LNVFLAIAVD NLAEAESLTS AQKAKAEERK RRKMSRGLPD KTEEEKSVMA KKLEQKPKGE GIPTTAKL K VDEFESNVNE VKDPYPSADF PGDDEEDEPE IPVSPRPRPL AELQLKEKAV PIPEASSFFI FSPTNKVRVL CHRIVNATW FTNFILLFIL LSSAALAAED PIRAESVRNQ ILGYFDIAFT SVFTVEIVLK MTTYGAFLHK GSFCRNYFNI LDLLVVAVSL ISMGLESST ISVVKILRVL RVLRPLRAIN RAKGLKHVVQ CVFVAIRTIG NIVLVTTLLQ FMFACIGVQL FKGKFFSCND L SKMTEEEC RGYYYVYKDG DPTQMELRPR QWIHNDFHFD NVLSAMMSLF TVSTFEGWPQ LLYRAIDSNE EDMGPVYNNR VE MAIFFII YIILIAFFMM NIFVGFVIVT FQEQGETEYK NCELDKNQRQ CVQYALKARP LRCYIPKNPY QYQVWYVVTS SYF EYLMFA LIMLNTICLG MQHYHQSEEM NHISDILNVA FTIIFTLEMI LKLLAFKARG YFGDPWNVFD FLIVIGSIID VILS EIDTF LASSGGLYCL GGGCGNVDPD ESARISSAFF RLFRVMRLIK LLSRAEGVRT LLWTFIKSFQ ALPYVALLIV MLFFI YAVI GMQMFGKIAL VDGTQINRNN NFQTFPQAVL LLFRCATGEA WQEILLACSY GKLCDPESDY APGEEYTCGT NFAYYY FIS FYMLCAFLII NLFVAVIMDN FDYLTRDWSI LGPHHLDEFK AIWAEYDPEA KGRIKHLDVV TLLRRIQPPL GFGKFCP HR VACKRLVGMN MPLNSDGTVT FNATLFALVR TALKIKTEGN FEQANEELRA IIKKIWKRTS MKLLDQVIPP IGDDEVTV G KFYATFLIQE HFRKFMKRQE EYYGYRPKKD TVQIQAGLRT IEEEAAPEIR RTISGDLTAE EELERAMVEA AMEERIFRR TGGLFGQVDT FLERTNSLPP VMANQRPLQF AEIEMEELES PVFLEDFPQD ARTNPLARAN TNNANANVAY GNSNHSNNQM FSSVHCERE FPGEAETPAA GRGALSHSHR ALGPHSKPCA GKLNGQLVQP GMPINQAPPA PCQQPSTDPP ERGQRRTSLT G SLQDEAPQ RRSSEGSTPR RPAPATALLI QEALVRGGLD TLAADAGFVT ATSQALADAC QMEPEEVEVA ATELLKARES VQ GMASVPG SLSRRSSLGS LDQVQGSQET LIPPRP UniProtKB: Voltage-dependent L-type calcium channel subunit alpha-1S |
-Macromolecule #2: Voltage-dependent calcium channel gamma-1 subunit
| Macromolecule | Name: Voltage-dependent calcium channel gamma-1 subunit / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.082254 KDa |
| Sequence | String: MSPTEAPKVR VTLFCILVGI VLAMTAVVSD HWAVLSPHME NHNTTCEAAH FGLWRICTKR IALGEDRSCG PITLPGEKNC SYFRHFNPG ESSEIFEFTT QKEYSISAAA ISVFSLGFLI MGTICALMAF RKKRDYLLRP ASMFYVFAGL CLFVSLEVMR Q SVKRMIDS ...String: MSPTEAPKVR VTLFCILVGI VLAMTAVVSD HWAVLSPHME NHNTTCEAAH FGLWRICTKR IALGEDRSCG PITLPGEKNC SYFRHFNPG ESSEIFEFTT QKEYSISAAA ISVFSLGFLI MGTICALMAF RKKRDYLLRP ASMFYVFAGL CLFVSLEVMR Q SVKRMIDS EDTVWIEYYY SWSFACACAA FVLLFLGGIS LLLFSLPRMP QNPWESCMDA EPEH UniProtKB: Voltage-dependent calcium channel gamma-1 subunit |
-Macromolecule #3: Voltage-dependent calcium channel subunit alpha-2/delta-1
| Macromolecule | Name: Voltage-dependent calcium channel subunit alpha-2/delta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 125.082906 KDa |
| Sequence | String: MAAGRPLAWT LTLWQAWLIL IGPSSEEPFP SAVTIKSWVD KMQEDLVTLA KTASGVHQLV DIYEKYQDLY TVEPNNARQL VEIAARDIE KLLSNRSKAL VRLALEAEKV QAAHQWREDF ASNEVVYYNA KDDLDPEKND SEPGSQRIKP VFIDDANFRR Q VSYQHAAV ...String: MAAGRPLAWT LTLWQAWLIL IGPSSEEPFP SAVTIKSWVD KMQEDLVTLA KTASGVHQLV DIYEKYQDLY TVEPNNARQL VEIAARDIE KLLSNRSKAL VRLALEAEKV QAAHQWREDF ASNEVVYYNA KDDLDPEKND SEPGSQRIKP VFIDDANFRR Q VSYQHAAV HIPTDIYEGS TIVLNELNWT SALDDVFKKN REEDPSLLWQ VFGSATGLAR YYPASPWVDN SRTPNKIDLY DV RRRPWYI QGAASPKDML ILVDVSGSVS GLTLKLIRTS VSEMLETLSD DDFVNVASFN SNAQDVSCFQ HLVQANVRNK KVL KDAVNN ITAKGITDYK KGFSFAFEQL LNYNVSRANC NKIIMLFTDG GEERAQEIFA KYNKDKKVRV FTFSVGQHNY DRGP IQWMA CENKGYYYEI PSIGAIRINT QEYLDVLGRP MVLAGDKAKQ VQWTNVYLDA LELGLVITGT LPVFNITGQF ENKTN LKNQ LILGVMGVDV SLEDIKRLTP RFTLCPNGYY FAIDPNGYVL LHPNLQPKPI GVGIPTINLR KRRPNVQNPK SQEPVT LDF LDAELENDIK VEIRNKMIDG ESGEKTFRTL VKSQDERYID KGNRTYTWTP VNGTDYSLAL VLPTYSFYYI KAKIEET IT QARYSETLKP DNFEESGYTF LAPRDYCSDL KPSDNNTEFL LNFNEFIDRK TPNNPSCNTD LINRVLLDAG FTNELVQN Y WSKQKNIKGV KARFVVTDGG ITRVYPKEAG ENWQENPETY EDSFYKRSLD NDNYVFTAPY FNKSGPGAYE SGIMVSKAV EIYIQGKLLK PAVVGIKIDV NSWIENFTKT SIRDPCAGPV CDCKRNSDVM DCVILDDGGF LLMANHDDYT NQIGRFFGEI DPSLMRHLV NISVYAFNKS YDYQSVCEPG AAPKQGAGHR SAYVPSIADI LQIGWWATAA AWSILQQFLL SLTFPRLLEA A DMEDDDFT ASMSKQSCIT EQTQYFFDND SKSFSGVLDC GNCSRIFHVE KLMNTNLIFI MVESKGTCPC DTRLLIQAEQ TS DGPDPCD MVKQPRYRKG PDVCFDNNVL EDYTDCGGVS GLNPSLWSII GIQFVLLWLV SGSRHCLL UniProtKB: Voltage-dependent calcium channel subunit alpha-2/delta-1 |
-Macromolecule #4: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 4 / Number of copies: 1 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #5: 1,2-Distearoyl-sn-glycerophosphoethanolamine
| Macromolecule | Name: 1,2-Distearoyl-sn-glycerophosphoethanolamine / type: ligand / ID: 5 / Number of copies: 9 / Formula: 3PE |
|---|---|
| Molecular weight | Theoretical: 748.065 Da |
| Chemical component information |  ChemComp-3PE: |
-Macromolecule #6: 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE
| Macromolecule | Name: 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE / type: ligand / ID: 6 / Number of copies: 1 / Formula: PC1 |
|---|---|
| Molecular weight | Theoretical: 790.145 Da |
| Chemical component information |  ChemComp-PC1: |
-Macromolecule #7: methyl (4~{R})-2,6-dimethyl-5-nitro-4-[2-(trifluoromethyl)phenyl]...
| Macromolecule | Name: methyl (4~{R})-2,6-dimethyl-5-nitro-4-[2-(trifluoromethyl)phenyl]-1,4-dihydropyridine-3-carboxylate type: ligand / ID: 7 / Number of copies: 1 / Formula: VFY |
|---|---|
| Molecular weight | Theoretical: 357.304 Da |
| Chemical component information | 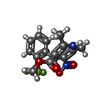 ChemComp-VFY: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 5 seconds before plunging. |
| Details | rCav1.1-100R was in lipid nanodisc |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















