+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7eb2 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human GABA(B) receptor-Gi protein complex | ||||||
 Components Components |
| ||||||
 Keywords Keywords | MEMBRANE PROTEIN / GABAB / Cryo-EM / GPCR / Gi | ||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of gamma-aminobutyric acid secretion / GABA B receptor activation / G protein-coupled GABA receptor complex / G protein-coupled neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential / neuron-glial cell signaling / G protein-coupled neurotransmitter receptor activity involved in regulation of presynaptic membrane potential / negative regulation of dopamine secretion / G protein-coupled GABA receptor activity / G protein-coupled receptor heterodimeric complex / negative regulation of epinephrine secretion ...negative regulation of gamma-aminobutyric acid secretion / GABA B receptor activation / G protein-coupled GABA receptor complex / G protein-coupled neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential / neuron-glial cell signaling / G protein-coupled neurotransmitter receptor activity involved in regulation of presynaptic membrane potential / negative regulation of dopamine secretion / G protein-coupled GABA receptor activity / G protein-coupled receptor heterodimeric complex / negative regulation of epinephrine secretion / positive regulation of growth hormone secretion / extracellular matrix protein binding / GABA receptor complex / negative regulation of adenylate cyclase activity / Class C/3 (Metabotropic glutamate/pheromone receptors) / negative regulation of synaptic transmission / positive regulation of glutamate secretion / gamma-aminobutyric acid signaling pathway / synaptic transmission, GABAergic / axolemma / adenylate cyclase inhibitor activity / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / D2 dopamine receptor binding / response to prostaglandin E / adenylate cyclase regulator activity / G protein-coupled serotonin receptor binding / adenylate cyclase-inhibiting serotonin receptor signaling pathway / cellular response to forskolin / regulation of mitotic spindle organization / dendritic shaft / response to nicotine / Regulation of insulin secretion / positive regulation of cholesterol biosynthetic process / negative regulation of insulin secretion / G protein-coupled receptor binding / response to peptide hormone / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / GABA-ergic synapse / mitochondrial membrane / Schaffer collateral - CA1 synapse / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through CDC42 / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / osteoblast differentiation / ADP signalling through P2Y purinoceptor 12 / synaptic vesicle / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / GDP binding / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / cellular response to catecholamine stimulus / transmembrane signaling receptor activity / ADORA2B mediated anti-inflammatory cytokines production / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / adenylate cyclase-activating dopamine receptor signaling pathway / GPER1 signaling / Inactivation, recovery and regulation of the phototransduction cascade / cellular response to prostaglandin E stimulus / G-protein beta-subunit binding / heterotrimeric G-protein complex / G alpha (12/13) signalling events / sensory perception of taste / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye / G protein activity / presynaptic membrane / GTPase binding / Ca2+ pathway / fibroblast proliferation / response to ethanol / midbody / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / cell cortex / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human)synthetic construct (others) | ||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.5 Å | ||||||
 Authors Authors | Shen, C. / Mao, C. / Xu, C. / Jin, N. / Zhang, H. / Shen, D. / Shen, Q. / Wang, X. / Hou, T. / Rondard, P. ...Shen, C. / Mao, C. / Xu, C. / Jin, N. / Zhang, H. / Shen, D. / Shen, Q. / Wang, X. / Hou, T. / Rondard, P. / Chen, Z. / Pin, J. / Zhang, Y. / Liu, J. | ||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: Structural basis of GABA receptor-G protein coupling. Authors: Cangsong Shen / Chunyou Mao / Chanjuan Xu / Nan Jin / Huibing Zhang / Dan-Dan Shen / Qingya Shen / Xiaomei Wang / Tingjun Hou / Zhong Chen / Philippe Rondard / Jean-Philippe Pin / Yan Zhang / Jianfeng Liu /   Abstract: G-protein-coupled receptors (GPCRs) have central roles in intercellular communication. Structural studies have revealed how GPCRs can activate G proteins. However, whether this mechanism is ...G-protein-coupled receptors (GPCRs) have central roles in intercellular communication. Structural studies have revealed how GPCRs can activate G proteins. However, whether this mechanism is conserved among all classes of GPCR remains unknown. Here we report the structure of the class-C heterodimeric GABA receptor, which is activated by the inhibitory transmitter GABA, in its active form complexed with G protein. We found that a single G protein interacts with the GB2 subunit of the GABA receptor at a site that mainly involves intracellular loop 2 on the side of the transmembrane domain. This is in contrast to the G protein binding in a central cavity, as has been observed with other classes of GPCR. This binding mode results from the active form of the transmembrane domain of this GABA receptor being different from that of other GPCRs, as it shows no outside movement of transmembrane helix 6. Our work also provides details of the inter- and intra-subunit changes that link agonist binding to G-protein activation in this heterodimeric complex. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7eb2.cif.gz 7eb2.cif.gz | 431.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7eb2.ent.gz pdb7eb2.ent.gz | 337.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7eb2.json.gz 7eb2.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  7eb2_validation.pdf.gz 7eb2_validation.pdf.gz | 875.3 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  7eb2_full_validation.pdf.gz 7eb2_full_validation.pdf.gz | 895.2 KB | Display | |
| Data in XML |  7eb2_validation.xml.gz 7eb2_validation.xml.gz | 63.9 KB | Display | |
| Data in CIF |  7eb2_validation.cif.gz 7eb2_validation.cif.gz | 97.1 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/eb/7eb2 https://data.pdbj.org/pub/pdb/validation_reports/eb/7eb2 ftp://data.pdbj.org/pub/pdb/validation_reports/eb/7eb2 ftp://data.pdbj.org/pub/pdb/validation_reports/eb/7eb2 | HTTPS FTP |
-Related structure data
| Related structure data |  31049MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Guanine nucleotide-binding protein ... , 3 types, 3 molecules ABY
| #1: Protein | Mass: 40414.047 Da / Num. of mol.: 1 / Mutation: S47N,G203A,E245A,A326S Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GNAI1 / Cell (production host): Hi5 / Production host: Homo sapiens (human) / Gene: GNAI1 / Cell (production host): Hi5 / Production host:  Trichoplusia ni (cabbage looper) / References: UniProt: P63096 Trichoplusia ni (cabbage looper) / References: UniProt: P63096 |
|---|---|
| #2: Protein | Mass: 39418.086 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GNB1 / Cell (production host): Hi5 / Production host: Homo sapiens (human) / Gene: GNB1 / Cell (production host): Hi5 / Production host:  Trichoplusia ni (cabbage looper) / References: UniProt: P62873 Trichoplusia ni (cabbage looper) / References: UniProt: P62873 |
| #3: Protein | Mass: 7861.143 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GNG2 / Cell (production host): Hi5 / Production host: Homo sapiens (human) / Gene: GNG2 / Cell (production host): Hi5 / Production host:  Trichoplusia ni (cabbage looper) / References: UniProt: P59768 Trichoplusia ni (cabbage looper) / References: UniProt: P59768 |
-Gamma-aminobutyric acid type B receptor subunit ... , 2 types, 2 molecules CD
| #5: Protein | Mass: 105976.297 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GABBR1, GPRC3A / Cell line (production host): HEK 293F / Production host: Homo sapiens (human) / Gene: GABBR1, GPRC3A / Cell line (production host): HEK 293F / Production host:  Homo sapiens (human) / References: UniProt: Q9UBS5 Homo sapiens (human) / References: UniProt: Q9UBS5 |
|---|---|
| #6: Protein | Mass: 91359.445 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GABBR2, GPR51, GPRC3B / Cell line (production host): HEK 293F / Production host: Homo sapiens (human) / Gene: GABBR2, GPR51, GPRC3B / Cell line (production host): HEK 293F / Production host:  Homo sapiens (human) / References: UniProt: O75899 Homo sapiens (human) / References: UniProt: O75899 |
-Antibody , 1 types, 1 molecules N
| #4: Antibody | Mass: 28813.047 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.) synthetic construct (others) / Cell (production host): Hi5 / Production host:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
|---|
-Non-polymers , 2 types, 2 molecules 
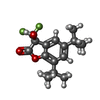

| #7: Chemical | ChemComp-2C0 / |
|---|---|
| #8: Chemical | ChemComp-FN0 / ( |
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component |
| ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source (natural) |
| ||||||||||||||||||||||||||||||||
| Source (recombinant) |
| ||||||||||||||||||||||||||||||||
| Buffer solution | pH: 7.5 | ||||||||||||||||||||||||||||||||
| Buffer component |
| ||||||||||||||||||||||||||||||||
| Specimen | Conc.: 1.8 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | ||||||||||||||||||||||||||||||||
| Specimen support | Grid material: GOLD / Grid mesh size: 300 divisions/in. / Grid type: Quantifoil R1.2/1.3 | ||||||||||||||||||||||||||||||||
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 29000 X / Calibrated magnification: 49310 X / Cs: 2.7 mm |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Electron dose: 64 e/Å2 / Detector mode: COUNTING / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Num. of real images: 13843 |
| Image scans | Movie frames/image: 40 / Used frames/image: 1-40 |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.18_3845: / Classification: refinement | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||||||
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 5889932 | ||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.5 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 275089 / Symmetry type: POINT | ||||||||||||||||||||||||||||
| Atomic model building |
| ||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller










 PDBj
PDBj


































