[English] 日本語
 Yorodumi
Yorodumi- PDB-7dkz: Structure of plant photosystem I-light harvesting complex I super... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7dkz | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of plant photosystem I-light harvesting complex I supercomplex | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | PHOTOSYNTHESIS / PSI-LHCI high plants | |||||||||
| Function / homology |  Function and homology information Function and homology informationphotosynthesis, light harvesting / chloroplast thylakoid lumen / photosystem I reaction center / photosystem I / photosynthetic electron transport in photosystem I / photosystem I / photosystem II / chlorophyll binding / chloroplast thylakoid membrane / photosynthesis ...photosynthesis, light harvesting / chloroplast thylakoid lumen / photosystem I reaction center / photosystem I / photosynthetic electron transport in photosystem I / photosystem I / photosystem II / chlorophyll binding / chloroplast thylakoid membrane / photosynthesis / chloroplast / 4 iron, 4 sulfur cluster binding / electron transfer activity / oxidoreductase activity / magnesium ion binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.393 Å MOLECULAR REPLACEMENT / Resolution: 2.393 Å | |||||||||
 Authors Authors | Wang, J. / Yu, L.J. / Wang, W. | |||||||||
 Citation Citation |  Journal: J Integr Plant Biol / Year: 2021 Journal: J Integr Plant Biol / Year: 2021Title: Structure of plant photosystem I-light harvesting complex I supercomplex at 2.4 angstrom resolution. Authors: Wang, J. / Yu, L.J. / Wang, W. / Yan, Q. / Kuang, T. / Qin, X. / Shen, J.R. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7dkz.cif.gz 7dkz.cif.gz | 1.8 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7dkz.ent.gz pdb7dkz.ent.gz | 1.5 MB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7dkz.json.gz 7dkz.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  7dkz_validation.pdf.gz 7dkz_validation.pdf.gz | 48 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  7dkz_full_validation.pdf.gz 7dkz_full_validation.pdf.gz | 47.8 MB | Display | |
| Data in XML |  7dkz_validation.xml.gz 7dkz_validation.xml.gz | 217.3 KB | Display | |
| Data in CIF |  7dkz_validation.cif.gz 7dkz_validation.cif.gz | 254 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/dk/7dkz https://data.pdbj.org/pub/pdb/validation_reports/dk/7dkz ftp://data.pdbj.org/pub/pdb/validation_reports/dk/7dkz ftp://data.pdbj.org/pub/pdb/validation_reports/dk/7dkz | HTTPS FTP |
-Related structure data
| Related structure data |  4xk8S S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 10 types, 10 molecules 12CDEFGHLK
| #1: Protein | Mass: 21477.596 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|---|
| #2: Protein | Mass: 28912.719 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
| #7: Protein | Mass: 8991.474 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
| #8: Protein | Mass: 16041.408 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
| #9: Protein | Mass: 7479.422 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
| #10: Protein | Mass: 17238.000 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
| #11: Protein | Mass: 10678.027 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
| #12: Protein | Mass: 9491.742 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
| #15: Protein | Mass: 16564.977 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
| #16: Protein | Mass: 7988.296 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
-Chlorophyll a-b binding protein ... , 2 types, 2 molecules 34
| #3: Protein | Mass: 29634.801 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|---|
| #4: Protein | Mass: 21994.029 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
-Photosystem I P700 chlorophyll a apoprotein ... , 2 types, 2 molecules AB
| #5: Protein | Mass: 84279.562 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|---|
| #6: Protein | Mass: 82512.938 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
-Photosystem I reaction center subunit ... , 2 types, 2 molecules IJ
| #13: Protein/peptide | Mass: 4474.527 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|---|
| #14: Protein/peptide | Mass: 4767.609 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
-Sugars , 3 types, 12 molecules 
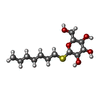



| #23: Sugar | | #25: Sugar | ChemComp-HTG / #26: Sugar | |
|---|
-Non-polymers , 10 types, 296 molecules 


















| #17: Chemical | ChemComp-CLA / #18: Chemical | ChemComp-CHL / #19: Chemical | ChemComp-LUT / ( #20: Chemical | ChemComp-XAT / ( #21: Chemical | ChemComp-BCR / #22: Chemical | ChemComp-LHG / #24: Chemical | ChemComp-LMG / #27: Chemical | #28: Chemical | #29: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 4.05 Å3/Da / Density % sol: 69.59 % |
|---|---|
| Crystal grow | Temperature: 285 K / Method: vapor diffusion, sitting drop / pH: 9 / Details: sodium chlotide, magnesium chloride, PEG 400 / PH range: 8.0-9.0 |
-Data collection
| Diffraction | Mean temperature: 80 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRF SSRF  / Beamline: BL17U / Wavelength: 0.979 Å / Beamline: BL17U / Wavelength: 0.979 Å |
| Detector | Type: ADSC QUANTUM 315r / Detector: CCD / Date: Jul 1, 2018 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.979 Å / Relative weight: 1 |
| Reflection | Resolution: 2.39→44.41 Å / Num. obs: 230250 / % possible obs: 98.84 % / Redundancy: 6.8 % / Biso Wilson estimate: 65.37 Å2 / CC1/2: 0.998 / Rpim(I) all: 0.04 / Rrim(I) all: 0.11 / Net I/σ(I): 17.8 |
| Reflection shell | Resolution: 2.39→2.48 Å / Redundancy: 6.2 % / Num. unique obs: 21565 / CC1/2: 0.708 / % possible all: 92.9 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 4XK8 Resolution: 2.393→44.405 Å / SU ML: 0.29 / Cross valid method: THROUGHOUT / σ(F): 1.35 / Phase error: 27.23 / Stereochemistry target values: ML
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 307.68 Å2 / Biso mean: 101.2191 Å2 / Biso min: 30 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 2.393→44.405 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Rfactor Rfree error: 0
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Origin x: 33.6643 Å / Origin y: 66.1507 Å / Origin z: 28.6623 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj





















