[English] 日本語
 Yorodumi
Yorodumi- PDB-7acw: LID/HID (LMW SLP and HMW SLP interacting domains) from C. diffici... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7acw | ||||||
|---|---|---|---|---|---|---|---|
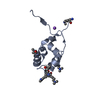
| Title | LID/HID (LMW SLP and HMW SLP interacting domains) from C. difficile (R7404 strain) | ||||||
 Components Components | (S-layer protein) x 2 | ||||||
 Keywords Keywords | STRUCTURAL PROTEIN / Bacterial surface / S-layer | ||||||
| Function / homology | Low molecular weight S layer protein, N-terminal / Low molecular weight S layer protein, N-terminal, subdomain / Low molecular weight S layer protein N terminal / : / Putative cell wall binding repeat 2 / Cell wall binding domain 2 (CWB2) / N-acetylmuramoyl-L-alanine amidase activity / S-layer protein Function and homology information Function and homology information | ||||||
| Biological species |  Clostridioides difficile (bacteria) Clostridioides difficile (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / AB INITIO PHASING / Resolution: 1.5 Å SYNCHROTRON / AB INITIO PHASING / Resolution: 1.5 Å | ||||||
 Authors Authors | Barwinska-Sendra, A. / Lanzoni-Mangutchi, P. / Basle, A. / Salgado, P.S. | ||||||
| Funding support |  United Kingdom, 1items United Kingdom, 1items
| ||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structure and assembly of the S-layer in C. difficile. Authors: Paola Lanzoni-Mangutchi / Oishik Banerji / Jason Wilson / Anna Barwinska-Sendra / Joseph A Kirk / Filipa Vaz / Shauna O'Beirne / Arnaud Baslé / Kamel El Omari / Armin Wagner / Neil F ...Authors: Paola Lanzoni-Mangutchi / Oishik Banerji / Jason Wilson / Anna Barwinska-Sendra / Joseph A Kirk / Filipa Vaz / Shauna O'Beirne / Arnaud Baslé / Kamel El Omari / Armin Wagner / Neil F Fairweather / Gillian R Douce / Per A Bullough / Robert P Fagan / Paula S Salgado /   Abstract: Many bacteria and archaea possess a two-dimensional protein array, or S-layer, that covers the cell surface and plays crucial roles in cell physiology. Here, we report the crystal structure of SlpA, ...Many bacteria and archaea possess a two-dimensional protein array, or S-layer, that covers the cell surface and plays crucial roles in cell physiology. Here, we report the crystal structure of SlpA, the main S-layer protein of the bacterial pathogen Clostridioides difficile, and use electron microscopy to study S-layer organisation and assembly. The SlpA crystal lattice mimics S-layer assembly in the cell, through tiling of triangular prisms above the cell wall, interlocked by distinct ridges facing the environment. Strikingly, the array is very compact, with pores of only ~10 Å in diameter, compared to other S-layers (30-100 Å). The surface-exposed flexible ridges are partially dispensable for overall structure and assembly, although a mutant lacking this region becomes susceptible to lysozyme, an important molecule in host defence. Thus, our work gives insights into S-layer organisation and provides a basis for development of C. difficile-specific therapeutics. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7acw.cif.gz 7acw.cif.gz | 102.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7acw.ent.gz pdb7acw.ent.gz | 78.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7acw.json.gz 7acw.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ac/7acw https://data.pdbj.org/pub/pdb/validation_reports/ac/7acw ftp://data.pdbj.org/pub/pdb/validation_reports/ac/7acw ftp://data.pdbj.org/pub/pdb/validation_reports/ac/7acw | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein | Mass: 8727.751 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Details: RT017, SLCT7b / Source: (gene. exp.)  Clostridioides difficile (bacteria) / Gene: slpA, SAMEA708418_00075 / Production host: Clostridioides difficile (bacteria) / Gene: slpA, SAMEA708418_00075 / Production host:  References: UniProt: Q9AEM2, N-acetylmuramoyl-L-alanine amidase #2: Protein/peptide | Mass: 5766.458 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Details: RT017, SLCT7b / Source: (gene. exp.)  Clostridioides difficile (bacteria) / Gene: slpA, SAMEA708418_00075 / Production host: Clostridioides difficile (bacteria) / Gene: slpA, SAMEA708418_00075 / Production host:  References: UniProt: Q9AEM2, N-acetylmuramoyl-L-alanine amidase #3: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.86 Å3/Da / Density % sol: 34 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 6.5 / Details: 1.6M sodium citrate tribasic dihidrate |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I04 / Wavelength: 0.9795 Å / Beamline: I04 / Wavelength: 0.9795 Å |
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Oct 18, 2018 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9795 Å / Relative weight: 1 |
| Reflection | Resolution: 1.5→52.01 Å / Num. obs: 34063 / % possible obs: 98.9 % / Redundancy: 3.4 % / Biso Wilson estimate: 22.6 Å2 / CC1/2: 0.998 / CC star: 0.999 / Rmerge(I) obs: 0.047 / Rpim(I) all: 0.028 / Rrim(I) all: 0.055 / Net I/σ(I): 8.1 |
| Reflection shell | Resolution: 1.5→1.55 Å / Redundancy: 2.6 % / Rmerge(I) obs: 0.22 / Num. unique obs: 3318 / CC1/2: 0.975 / CC star: 0.994 / Rpim(I) all: 0.154 / Rrim(I) all: 0.27 / % possible all: 95.3 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure: AB INITIO PHASING / Resolution: 1.5→52.01 Å / Cor.coef. Fo:Fc: 0.973 / Cor.coef. Fo:Fc free: 0.961 / SU B: 4.506 / SU ML: 0.07 / Cross valid method: THROUGHOUT / ESU R: 0.088 / ESU R Free: 0.076 / Stereochemistry target values: MAXIMUM LIKELIHOOD Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS U VALUES : REFINED INDIVIDUALLY
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.7 Å / Shrinkage radii: 0.7 Å / VDW probe radii: 1.3 Å / Solvent model: MASK | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 77.08 Å2 / Biso mean: 32.518 Å2 / Biso min: 20.98 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 1.5→52.01 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.5→1.539 Å / Rfactor Rfree error: 0 / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller

















 PDBj
PDBj
