[English] 日本語
 Yorodumi
Yorodumi- PDB-6yjf: Plasmoodium vivax phosphoglycerate kinase bound to nitrofuran inh... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6yjf | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Plasmoodium vivax phosphoglycerate kinase bound to nitrofuran inhibitor from PEGSmear at pH 6.5 | |||||||||
 Components Components | Phosphoglycerate kinase | |||||||||
 Keywords Keywords | BIOSYNTHETIC PROTEIN / phosphoglycerate kinase / metabolic enzyme / kinase | |||||||||
| Function / homology |  Function and homology information Function and homology informationphosphoglycerate kinase / phosphoglycerate kinase activity / glycolytic process / gluconeogenesis / ADP binding / ATP binding / metal ion binding / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 1.85 Å molecular replacement / Resolution: 1.85 Å | |||||||||
 Authors Authors | Hyvonen, M. / Brear, P. / Blaszczyk, B.K. | |||||||||
 Citation Citation |  Journal: to be published Journal: to be publishedTitle: Phosphoglycerate Kinase as a potential target for antimalarial therapy Authors: King, L. / Brear, P. / Bourgard, C. / Cassiano, C. / Mota, D. / Blaszczyk, B.K. / Tomaz, K. / Khedim, M. / Furlan, M. / Ramos, P. / Andresen, E. / Tarczykowska, A. / Tiburcio, L. / Kurowska, ...Authors: King, L. / Brear, P. / Bourgard, C. / Cassiano, C. / Mota, D. / Blaszczyk, B.K. / Tomaz, K. / Khedim, M. / Furlan, M. / Ramos, P. / Andresen, E. / Tarczykowska, A. / Tiburcio, L. / Kurowska, A. / Sulskis, D. / Oliver, S. / Grotli, M. / Massirer, K. / Saphire, E. / Burmann, B. / Witkowski, B. / Costa, F. / Sunnerhagen, P. / Hyvonen, M. / Bilsland, E. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6yjf.cif.gz 6yjf.cif.gz | 100.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6yjf.ent.gz pdb6yjf.ent.gz | 74.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6yjf.json.gz 6yjf.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  6yjf_validation.pdf.gz 6yjf_validation.pdf.gz | 748.8 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  6yjf_full_validation.pdf.gz 6yjf_full_validation.pdf.gz | 749 KB | Display | |
| Data in XML |  6yjf_validation.xml.gz 6yjf_validation.xml.gz | 18.4 KB | Display | |
| Data in CIF |  6yjf_validation.cif.gz 6yjf_validation.cif.gz | 27.6 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/yj/6yjf https://data.pdbj.org/pub/pdb/validation_reports/yj/6yjf ftp://data.pdbj.org/pub/pdb/validation_reports/yj/6yjf ftp://data.pdbj.org/pub/pdb/validation_reports/yj/6yjf | HTTPS FTP |
-Related structure data
| Related structure data |  6y3aSC  6yjeC S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 45199.023 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Gene: PVC01_070025900, PVP01_0721000, PVT01_070026100 / Plasmid: pHAT2 / Production host:  |
|---|---|
| #2: Chemical | ChemComp-GOL / |
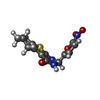
| #3: Chemical | ChemComp-OTQ / ( |
| #4: Water | ChemComp-HOH / |
| Has ligand of interest | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.41 Å3/Da / Density % sol: 49.05 % |
|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, sitting drop / pH: 6.5 / Details: 25 %v/v PEGSM, 0.1 M MES 6.5 pH |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I03 / Wavelength: 0.9762 Å / Beamline: I03 / Wavelength: 0.9762 Å | ||||||||||||||||||||||||||||||
| Detector | Type: DECTRIS EIGER2 XE 16M / Detector: PIXEL / Date: Mar 5, 2020 | ||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 0.9762 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||
| Reflection | Resolution: 1.85→68.66 Å / Num. obs: 34744 / % possible obs: 91.3 % / Redundancy: 12.9 % / Biso Wilson estimate: 38.35 Å2 / CC1/2: 0.999 / Rmerge(I) obs: 0.094 / Rpim(I) all: 0.027 / Rrim(I) all: 0.098 / Net I/σ(I): 14.3 / Num. measured all: 448345 | ||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
-Phasing
| Phasing | Method:  molecular replacement molecular replacement |
|---|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 6Y3A Resolution: 1.85→25.05 Å / Cor.coef. Fo:Fc: 0.935 / Cor.coef. Fo:Fc free: 0.913 / SU R Cruickshank DPI: 0.152 / Cross valid method: THROUGHOUT / σ(F): 0 / SU R Blow DPI: 0.16 / SU Rfree Blow DPI: 0.141 / SU Rfree Cruickshank DPI: 0.138
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 125.09 Å2 / Biso mean: 45.79 Å2 / Biso min: 27.17 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 1.85→25.05 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.85→1.91 Å / Rfactor Rfree error: 0 / Total num. of bins used: 17
|
 Movie
Movie Controller
Controller












 PDBj
PDBj