[English] 日本語
 Yorodumi
Yorodumi- PDB-6vjv: Crystal structure of the Prochlorococcus phage (myovirus P-SSM2) ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6vjv | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure of the Prochlorococcus phage (myovirus P-SSM2) ferredoxin at 1.6 Angstroms | ||||||||||||
 Components Components | Ferredoxin | ||||||||||||
 Keywords Keywords | ELECTRON TRANSPORT / Iron Sulfur Cluster Binding / myovirus P-SSM2 / Prochlorococcus phage / ferredoxin / 2 Iron 2 Sulfur | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationelectron transport chain / 2 iron, 2 sulfur cluster binding / electron transfer activity / metal ion binding Similarity search - Function | ||||||||||||
| Biological species |  Prochlorococcus phage P-SSM2 (virus) Prochlorococcus phage P-SSM2 (virus) | ||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.59 Å MOLECULAR REPLACEMENT / Resolution: 1.59 Å | ||||||||||||
 Authors Authors | Olmos Jr., J.L. / Campbell, I.J. / Miller, M.D. / Xu, W. / Kahanda, D. / Atkinson, J.T. / Sparks, N. / Bennett, G.N. / Silberg, J.J. / Phillips Jr., G.N. | ||||||||||||
| Funding support |  United States, 3items United States, 3items
| ||||||||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2020 Journal: J.Biol.Chem. / Year: 2020Title: Prochlorococcusphage ferredoxin: structural characterization and electron transfer to cyanobacterial sulfite reductases. Authors: Campbell, I.J. / Olmos Jr., J.L. / Xu, W. / Kahanda, D. / Atkinson, J.T. / Sparks, O.N. / Miller, M.D. / Phillips Jr., G.N. / Bennett, G.N. / Silberg, J.J. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6vjv.cif.gz 6vjv.cif.gz | 131.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6vjv.ent.gz pdb6vjv.ent.gz | 102.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6vjv.json.gz 6vjv.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  6vjv_validation.pdf.gz 6vjv_validation.pdf.gz | 305.4 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  6vjv_full_validation.pdf.gz 6vjv_full_validation.pdf.gz | 305.4 KB | Display | |
| Data in XML |  6vjv_validation.xml.gz 6vjv_validation.xml.gz | 1.5 KB | Display | |
| Data in CIF |  6vjv_validation.cif.gz 6vjv_validation.cif.gz | 4.8 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/vj/6vjv https://data.pdbj.org/pub/pdb/validation_reports/vj/6vjv ftp://data.pdbj.org/pub/pdb/validation_reports/vj/6vjv ftp://data.pdbj.org/pub/pdb/validation_reports/vj/6vjv | HTTPS FTP |
-Related structure data
| Related structure data |  5h57S S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||
| 2 | 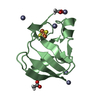
| ||||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 10381.252 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Prochlorococcus phage P-SSM2 (virus) / Gene: PCMG_00283, PSSM2_281 / Production host: Prochlorococcus phage P-SSM2 (virus) / Gene: PCMG_00283, PSSM2_281 / Production host:  #2: Chemical | #3: Chemical | ChemComp-ACT / #4: Chemical | ChemComp-ZN / #5: Water | ChemComp-HOH / | Has ligand of interest | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.44 Å3/Da / Density % sol: 49.61 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 4.5 Details: 10% (w/v) PEG 3000, 200 mM zinc acetate, and 100 mM sodium acetate/acetic acid pH 4.5 |
-Data collection
| Diffraction | Mean temperature: 100 K / Ambient temp details: cryo-stream / Serial crystal experiment: N | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 23-ID-B / Wavelength: 1.033 Å / Beamline: 23-ID-B / Wavelength: 1.033 Å | |||||||||||||||||||||||||||
| Detector | Type: DECTRIS EIGER X 16M / Detector: PIXEL / Date: Feb 7, 2019 / Details: Adjustable focus K-B pair | |||||||||||||||||||||||||||
| Radiation | Monochromator: Si(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 1.033 Å / Relative weight: 1 | |||||||||||||||||||||||||||
| Reflection | Resolution: 1.589→52.57 Å / Num. obs: 27020 / % possible obs: 98.5 % / Redundancy: 6.6 % / CC1/2: 0.996 / Rmerge(I) obs: 0.12 / Rpim(I) all: 0.05 / Rrim(I) all: 0.131 / Net I/σ(I): 8.9 | |||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 5H57 Resolution: 1.59→39.06 Å / SU ML: 0.2 / Cross valid method: THROUGHOUT / σ(F): 1.33 / Phase error: 22.12 Details: In chain B, the iron-sulfur cluster site was disrupted. The density surrounding the cluster in chain B is consistent with a mixture of zinc bound and an intact 2Fe-2S cluster. To account for ...Details: In chain B, the iron-sulfur cluster site was disrupted. The density surrounding the cluster in chain B is consistent with a mixture of zinc bound and an intact 2Fe-2S cluster. To account for the poor density and the anomalous data, chain B has been modeled as a grouped occupancy, where the model is partially comprised of intact cysteine-coordinated iron-sulfur cluster, and a zinc ion with a hydration shell.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 108.9 Å2 / Biso mean: 33.7873 Å2 / Biso min: 11.18 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 1.59→39.06 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Rfactor Rfree error: 0 / Total num. of bins used: 9
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj















