+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6ug0 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | N2-bound Nitrogenase MoFe-protein from Azotobacter vinelandii | |||||||||
 Components Components | (Nitrogenase molybdenum-iron protein ...) x 2 | |||||||||
 Keywords Keywords | OXIDOREDUCTASE / Azotobacter vinelandii / MoFe-protein / Fe-protein / FeMo-cofactor / oxidized P-cluster | |||||||||
| Function / homology |  Function and homology information Function and homology informationmolybdenum-iron nitrogenase complex / nitrogenase / nitrogenase activity / nitrogen fixation / iron-sulfur cluster binding / ATP binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Azotobacter vinelandii (bacteria) Azotobacter vinelandii (bacteria) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.83 Å MOLECULAR REPLACEMENT / Resolution: 1.83 Å | |||||||||
 Authors Authors | Kang, W. / Hu, Y. / Ribbe, M.W. | |||||||||
| Funding support |  United States, 2items United States, 2items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2020 Journal: Science / Year: 2020Title: Structural evidence for a dynamic metallocofactor during N2reduction by Mo-nitrogenase. Authors: Kang, W. / Lee, C.C. / Jasniewski, A.J. / Ribbe, M.W. / Hu, Y. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6ug0.cif.gz 6ug0.cif.gz | 985.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6ug0.ent.gz pdb6ug0.ent.gz | 678.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6ug0.json.gz 6ug0.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ug/6ug0 https://data.pdbj.org/pub/pdb/validation_reports/ug/6ug0 ftp://data.pdbj.org/pub/pdb/validation_reports/ug/6ug0 ftp://data.pdbj.org/pub/pdb/validation_reports/ug/6ug0 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6vxtC  3u7qS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||
| Unit cell |
|
- Components
Components
-Nitrogenase molybdenum-iron protein ... , 2 types, 4 molecules ACBD
| #1: Protein | Mass: 55363.043 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Azotobacter vinelandii (bacteria) / Gene: nifD / Production host: Azotobacter vinelandii (bacteria) / Gene: nifD / Production host:  Azotobacter vinelandii (bacteria) / References: UniProt: P07328, nitrogenase Azotobacter vinelandii (bacteria) / References: UniProt: P07328, nitrogenase#2: Protein | Mass: 59535.879 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Azotobacter vinelandii (bacteria) / Gene: nifK / Production host: Azotobacter vinelandii (bacteria) / Gene: nifK / Production host:  Azotobacter vinelandii (bacteria) / References: UniProt: P07329, nitrogenase Azotobacter vinelandii (bacteria) / References: UniProt: P07329, nitrogenase |
|---|
-Non-polymers , 11 types, 809 molecules 









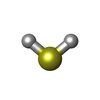





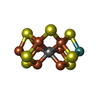

| #3: Chemical | | #4: Chemical | ChemComp-ICE / | #5: Chemical | #6: Chemical | ChemComp-H2S / #7: Chemical | ChemComp-MO / #8: Chemical | #9: Chemical | #10: Chemical | ChemComp-GOL / #11: Chemical | ChemComp-PGE / | #12: Chemical | ChemComp-ICZ / | #13: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.92 Å3/Da / Density % sol: 57.85 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 5 Details: 14-19 % w/v PEG smear high, 0.16M ammonium acetate, 0.1M sodium citrate (pH 5.0), 25.6 % v/v glycerol, 5mM Eu(II)-EGTA |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRL SSRL  / Beamline: BL12-2 / Wavelength: 1 Å / Beamline: BL12-2 / Wavelength: 1 Å |
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Jun 7, 2019 Details: Flat Si Rh coated M0, Kirkpatrick-Baez flat bent Si M1 & M2 |
| Radiation | Monochromator: Liquid nitrogen-cooled double crystal Si(111) Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 1.83→50 Å / Num. obs: 237283 / % possible obs: 95.2 % / Redundancy: 11.5 % / Biso Wilson estimate: 31.55 Å2 / CC1/2: 0.994 / Rpim(I) all: 0.061 / Rrim(I) all: 0.215 / Net I/σ(I): 11.5 |
| Reflection shell | Resolution: 1.83→1.86 Å / Redundancy: 9 % / Num. unique obs: 225235 / CC1/2: 0.352 / Rpim(I) all: 1.035 / Rrim(I) all: 3.163 / % possible all: 94.8 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB entry 3U7Q Resolution: 1.83→39.19 Å / SU ML: 0.3038 / Cross valid method: FREE R-VALUE / σ(F): 1.33 / Phase error: 32.6186 Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2 Details: Authors state that the electron density of metal clusters presented in validation report and wwPDB websites are calculated from general pipeline without applying specific geometry restraints ...Details: Authors state that the electron density of metal clusters presented in validation report and wwPDB websites are calculated from general pipeline without applying specific geometry restraints of the clusters. To reproduce accurate difference electron density map for the clusters, users should take the author-provided map coefficients presented in the structure factor file of the entry.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 49.87 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.83→39.19 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Origin x: 30.9250854555 Å / Origin y: 118.258636614 Å / Origin z: 151.142986479 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group | Selection details: all |
 Movie
Movie Controller
Controller























 PDBj
PDBj










