| Entry | Database: PDB / ID: 6vxt
|
|---|
| Title | Activated Nitrogenase MoFe-protein from Azotobacter vinelandii |
|---|
 Components Components | (Nitrogenase molybdenum-iron protein ...) x 2 |
|---|
 Keywords Keywords | OXIDOREDUCTASE / Azotobacter vinelandii / MoFe-protein / Fe-protein / FeMo-cofactor / P-cluster |
|---|
| Function / homology |  Function and homology information Function and homology information
molybdenum-iron nitrogenase complex / nitrogenase / nitrogenase activity / nitrogen fixation / iron-sulfur cluster binding / ATP binding / metal ion bindingSimilarity search - Function Nitrogenase Molybdenum-iron Protein, subunit B, domain 4 / Nitrogenase Molybdenum-iron Protein, subunit B; domain 4 / Nitrogenase molybdenum-iron protein beta chain, N-terminal / Domain of unknown function (DUF3364) / Nitrogenase molybdenum-iron protein alpha chain / Nitrogenase molybdenum-iron protein beta chain / Nitrogenase component 1, alpha chain / Nitrogenase component 1, conserved site / Nitrogenases component 1 alpha and beta subunits signature 2. / Nitrogenases component 1 alpha and beta subunits signature 1. ...Nitrogenase Molybdenum-iron Protein, subunit B, domain 4 / Nitrogenase Molybdenum-iron Protein, subunit B; domain 4 / Nitrogenase molybdenum-iron protein beta chain, N-terminal / Domain of unknown function (DUF3364) / Nitrogenase molybdenum-iron protein alpha chain / Nitrogenase molybdenum-iron protein beta chain / Nitrogenase component 1, alpha chain / Nitrogenase component 1, conserved site / Nitrogenases component 1 alpha and beta subunits signature 2. / Nitrogenases component 1 alpha and beta subunits signature 1. / : / Nitrogenase/oxidoreductase, component 1 / Nitrogenase component 1 type Oxidoreductase / Nitrogenase molybdenum iron protein domain / Up-down Bundle / Rossmann fold / 3-Layer(aba) Sandwich / Mainly Alpha / Alpha BetaSimilarity search - Domain/homology FE(8)-S(7) CLUSTER / : / HYDROSULFURIC ACID / 3-HYDROXY-3-CARBOXY-ADIPIC ACID / Chem-ICS / MOLYBDENUM ATOM / Nitrogenase molybdenum-iron protein alpha chain / Nitrogenase molybdenum-iron protein beta chainSimilarity search - Component |
|---|
| Biological species |  Azotobacter vinelandii (bacteria) Azotobacter vinelandii (bacteria) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.74 Å MOLECULAR REPLACEMENT / Resolution: 1.74 Å |
|---|
 Authors Authors | Kang, W. / Hu, Y. / Ribbe, M.W. |
|---|
| Funding support |  United States, 2items United States, 2items | Organization | Grant number | Country |
|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | GM67626 |  United States United States | | Department of Energy (DOE, United States) | DE-SC0016510 |  United States United States |
|
|---|
 Citation Citation |  Journal: Science / Year: 2020 Journal: Science / Year: 2020
Title: Structural evidence for a dynamic metallocofactor during N2reduction by Mo-nitrogenase.
Authors: Kang, W. / Lee, C.C. / Jasniewski, A.J. / Ribbe, M.W. / Hu, Y. |
|---|
| History | | Deposition | Feb 24, 2020 | Deposition site: RCSB / Processing site: RCSB |
|---|
| Revision 1.0 | Jun 24, 2020 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Jul 1, 2020 | Group: Database references / Category: citation / citation_author
Item: _citation.journal_volume / _citation.page_first ..._citation.journal_volume / _citation.page_first / _citation.page_last / _citation.pdbx_database_id_PubMed / _citation.title / _citation_author.identifier_ORCID |
|---|
| Revision 1.2 | Oct 11, 2023 | Group: Data collection / Database references ...Data collection / Database references / Derived calculations / Refinement description
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / pdbx_struct_conn_angle / struct_conn
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession ..._database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_struct_conn_angle.ptnr1_auth_asym_id / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr2_auth_asym_id / _pdbx_struct_conn_angle.ptnr2_auth_comp_id / _pdbx_struct_conn_angle.ptnr2_auth_seq_id / _pdbx_struct_conn_angle.ptnr2_label_asym_id / _pdbx_struct_conn_angle.ptnr2_label_atom_id / _pdbx_struct_conn_angle.ptnr2_label_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_asym_id / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.value / _struct_conn.pdbx_dist_value / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id |
|---|
|
|---|
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Azotobacter vinelandii (bacteria)
Azotobacter vinelandii (bacteria) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.74 Å
MOLECULAR REPLACEMENT / Resolution: 1.74 Å  Authors
Authors United States, 2items
United States, 2items  Citation
Citation Journal: Science / Year: 2020
Journal: Science / Year: 2020 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6vxt.cif.gz
6vxt.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6vxt.ent.gz
pdb6vxt.ent.gz PDB format
PDB format 6vxt.json.gz
6vxt.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/vx/6vxt
https://data.pdbj.org/pub/pdb/validation_reports/vx/6vxt ftp://data.pdbj.org/pub/pdb/validation_reports/vx/6vxt
ftp://data.pdbj.org/pub/pdb/validation_reports/vx/6vxt

 Links
Links Assembly
Assembly
 Components
Components Azotobacter vinelandii (bacteria) / References: UniProt: P07328, nitrogenase
Azotobacter vinelandii (bacteria) / References: UniProt: P07328, nitrogenase Azotobacter vinelandii (bacteria) / References: UniProt: P07329, nitrogenase
Azotobacter vinelandii (bacteria) / References: UniProt: P07329, nitrogenase
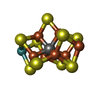





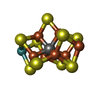





 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  SSRL
SSRL  / Beamline: BL12-2 / Wavelength: 1 Å
/ Beamline: BL12-2 / Wavelength: 1 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller




















 PDBj
PDBj






