| Entry | Database: PDB / ID: 6r3m
|
|---|
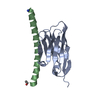
| Title | Family 11 Carbohydrate-Binding Module from Clostridium thermocellum in complex with beta-1,3-1,4-mixed-linked tetrasaccharide |
|---|
 Components Components | Endoglucanase H |
|---|
 Keywords Keywords | SUGAR BINDING PROTEIN / CtCBM11 beta-1 / 3-1 / 4-mixed-linked glucan Clostridium thermocellum |
|---|
| Function / homology |  Function and homology information Function and homology information
cellulase / cellulase activity / beta-glucosidase activity / cellulose catabolic process / cell surface / extracellular regionSimilarity search - Function Carbohydrate binding module family 11 / Carbohydrate binding domain (family 11) / Galactose-binding lectin / Glycosyl hydrolase family 26 / Glycosyl hydrolase family 26 domain / Glycosyl hydrolases family 26 (GH26) domain profile. / : / Clostridium cellulosome enzymes repeated domain signature. / Glycoside hydrolase, family 5, conserved site / Glycosyl hydrolases family 5 signature. ...Carbohydrate binding module family 11 / Carbohydrate binding domain (family 11) / Galactose-binding lectin / Glycosyl hydrolase family 26 / Glycosyl hydrolase family 26 domain / Glycosyl hydrolases family 26 (GH26) domain profile. / : / Clostridium cellulosome enzymes repeated domain signature. / Glycoside hydrolase, family 5, conserved site / Glycosyl hydrolases family 5 signature. / Dockerin domain / Dockerin domain profile. / Dockerin type I domain / Dockerin type I repeat / Dockerin domain superfamily / Glycoside hydrolase, family 5 / Cellulase (glycosyl hydrolase family 5) / Galactose-binding-like domain superfamily / EF-hand calcium-binding domain. / Glycoside hydrolase superfamily / Jelly Rolls / Sandwich / Mainly BetaSimilarity search - Domain/homology |
|---|
| Biological species |  Hungateiclostridium thermocellum ATCC 27405 (bacteria) Hungateiclostridium thermocellum ATCC 27405 (bacteria) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.45 Å MOLECULAR REPLACEMENT / Resolution: 1.45 Å |
|---|
 Authors Authors | Ribeiro, D.O. / Carvalho, A.L. |
|---|
| Funding support |  Portugal, 3items Portugal, 3items | Organization | Grant number | Country |
|---|
| Foundation for Science and Technology | SFRH/BD/100569/2014 |  Portugal Portugal | | Foundation for Science and Technology | PTDC/BBB-BEP/0869/2014 |  Portugal Portugal | | Foundation for Science and Technology | EXPL/IF/01621/2013 |  Portugal Portugal |
|
|---|
 Citation Citation |  Journal: Febs J. / Year: 2020 Journal: Febs J. / Year: 2020
Title: Molecular basis for the preferential recognition of beta 1,3-1,4-glucans by the family 11 carbohydrate-binding module from Clostridium thermocellum.
Authors: Ribeiro, D.O. / Viegas, A. / Pires, V.M.R. / Medeiros-Silva, J. / Bule, P. / Chai, W. / Marcelo, F. / Fontes, C.M.G.A. / Cabrita, E.J. / Palma, A.S. / Carvalho, A.L. |
|---|
| History | | Deposition | Mar 20, 2019 | Deposition site: PDBE / Processing site: PDBE |
|---|
| Revision 1.0 | Feb 5, 2020 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Jul 15, 2020 | Group: Database references / Category: citation / citation_author
Item: _citation.journal_volume / _citation.page_first ..._citation.journal_volume / _citation.page_first / _citation.page_last / _citation.year / _citation_author.identifier_ORCID |
|---|
| Revision 2.0 | Jul 29, 2020 | Group: Atomic model / Data collection ...Atomic model / Data collection / Derived calculations / Structure summary
Category: atom_site / chem_comp ...atom_site / chem_comp / entity / pdbx_branch_scheme / pdbx_chem_comp_identifier / pdbx_entity_branch / pdbx_entity_branch_descriptor / pdbx_entity_branch_link / pdbx_entity_branch_list / pdbx_entity_nonpoly / pdbx_nonpoly_scheme / pdbx_struct_assembly_gen / pdbx_struct_conn_angle / pdbx_struct_special_symmetry / struct_asym / struct_conn / struct_conn_type / struct_site / struct_site_gen
Item: _atom_site.B_iso_or_equiv / _atom_site.Cartn_x ..._atom_site.B_iso_or_equiv / _atom_site.Cartn_x / _atom_site.Cartn_y / _atom_site.Cartn_z / _atom_site.auth_asym_id / _atom_site.auth_atom_id / _atom_site.auth_comp_id / _atom_site.auth_seq_id / _atom_site.label_alt_id / _atom_site.label_asym_id / _atom_site.label_atom_id / _atom_site.label_comp_id / _atom_site.label_entity_id / _atom_site.occupancy / _atom_site.type_symbol / _chem_comp.name / _entity.formula_weight / _entity.pdbx_description / _entity.pdbx_number_of_molecules / _entity.src_method / _entity.type / _pdbx_struct_assembly_gen.asym_id_list / _pdbx_struct_conn_angle.ptnr2_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_special_symmetry.label_asym_id / _struct_conn.conn_type_id / _struct_conn.id / _struct_conn.pdbx_dist_value / _struct_conn.pdbx_leaving_atom_flag / _struct_conn.pdbx_ptnr1_label_alt_id / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn_type.id
Description: Carbohydrate remediation / Provider: repository / Type: Remediation |
|---|
| Revision 2.1 | Jan 24, 2024 | Group: Data collection / Database references ...Data collection / Database references / Refinement description / Structure summary
Category: chem_comp / chem_comp_atom ...chem_comp / chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model
Item: _chem_comp.pdbx_synonyms / _database_2.pdbx_DOI / _database_2.pdbx_database_accession |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Hungateiclostridium thermocellum ATCC 27405 (bacteria)
Hungateiclostridium thermocellum ATCC 27405 (bacteria) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.45 Å
MOLECULAR REPLACEMENT / Resolution: 1.45 Å  Authors
Authors Portugal, 3items
Portugal, 3items  Citation
Citation Journal: Febs J. / Year: 2020
Journal: Febs J. / Year: 2020 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6r3m.cif.gz
6r3m.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6r3m.ent.gz
pdb6r3m.ent.gz PDB format
PDB format 6r3m.json.gz
6r3m.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads 6r3m_validation.pdf.gz
6r3m_validation.pdf.gz wwPDB validaton report
wwPDB validaton report 6r3m_full_validation.pdf.gz
6r3m_full_validation.pdf.gz 6r3m_validation.xml.gz
6r3m_validation.xml.gz 6r3m_validation.cif.gz
6r3m_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/r3/6r3m
https://data.pdbj.org/pub/pdb/validation_reports/r3/6r3m ftp://data.pdbj.org/pub/pdb/validation_reports/r3/6r3m
ftp://data.pdbj.org/pub/pdb/validation_reports/r3/6r3m

 Links
Links Assembly
Assembly
 Components
Components Hungateiclostridium thermocellum ATCC 27405 (bacteria)
Hungateiclostridium thermocellum ATCC 27405 (bacteria)








 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  Diamond
Diamond  / Beamline: I02 / Wavelength: 0.9763 Å
/ Beamline: I02 / Wavelength: 0.9763 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller









 PDBj
PDBj




